[TOC]
###### tags: `comms`
# PAML Pilot
:::info
Anyone can edit this note.
Click on the pencil icon or open https://hackmd.io/@friendzymes/paml/edit.
Associated GitHub issue: https://github.com/friendzymes/community/issues/14.
:::
## 2021-12-16 Meet
### Agenda
- Sanity check and confirm/coordinate current protocol plan
- Set high-level OKRs and milestones.
- Bumped to Meeting 2
- Medium-term goal for BioProtocols: Get an alpha out by April 2022
- "LIMS-less interlab coordination"
### Notes
- Reviewing the protocols.io draft
- Steps 18 and 19 raise questions about how much to abstract
- One option is to explode "recovery" into multiple explicit steps
- Another option is to abstract "recovery" into another protocol for modularity
- Step 1
- 60 mass percent is suboptimal representation
- PAML config
- install flow needs work
- graphviz dependency isn't documented
- inconsistent results with pip/pip3
- PAML colab notebooks need plumbing as well
- Luiza notes on PAML_GG_protocol
- https://docs.google.com/document/d/1AupzE_OLUqp-3mfgJ47NplMxqvzMAK5I/edit#heading=h.gjdgxs
### To Discuss with BBN, etc.:
- Ontology is unclear for primitives.
- How are schemas for parameters resolved? When are they resolved?
- Are they expected to be stored in a "canonical" way in a central place like bioprotocols.org?
- If so, to what degree is it expected that people will "homebrew" their own?
- And also, we probably need to set that up, right?
- What is the intended difference in practice of a SampleArray and a SampleCollection?
- PAML spec questions
- Needed: exhaustive list of primitives
- Needed: list of recommended changes and additions
## 2021-12-04 Meet
### Recap
At COMBINE 2021, we heard about a new lab modeling language.
Jake Beal led an exploratory workshop on this spec.
Its description was as follows:
> **Defining a Common Protocol Representation**
> This session is a working session for stakeholders to discuss needs, requirements, and opportunities for a common protocol representation, leveraging the current protocol specification and implementation of the Protocol Activity Markup[sic] Language (PAML).
> [name=Jacob Beal] [time=Wed, Oct 13, 2021] [color=#0074D9]
This led to a planning session for preliminary working groups.
The aim of this second session was stated as follows:
> **Implementation and Roadmap for Common Protocol Representation**
> This session is a working session that will use the outcomes of the prior “Defining a Common Protocol Representation” session to work on the specifics of implementation of a common protocol representation, leveraging the current prototype specification and implementation of the Protocol Activity Markup[sic] Language (PAML), and to determine priorities and next steps.
> [time=Thu, Oct 14, 2021] [name=Jacob Beal] [color=#0074D9]
By the end of this session, the rough contours of three groups were defined.
1. GUI dev
1. iGEM interlab - iGEM engineering
1. Friendzymes and friends - practical pilot
Niches for which no group yet exists include:
1. Protocol dev
1. Core "data curation" - ontology dev, knowledge representation
1. Non-GUI toolchain dev
**We are here to initiate discussion of group 3: Friendzymes and friends**
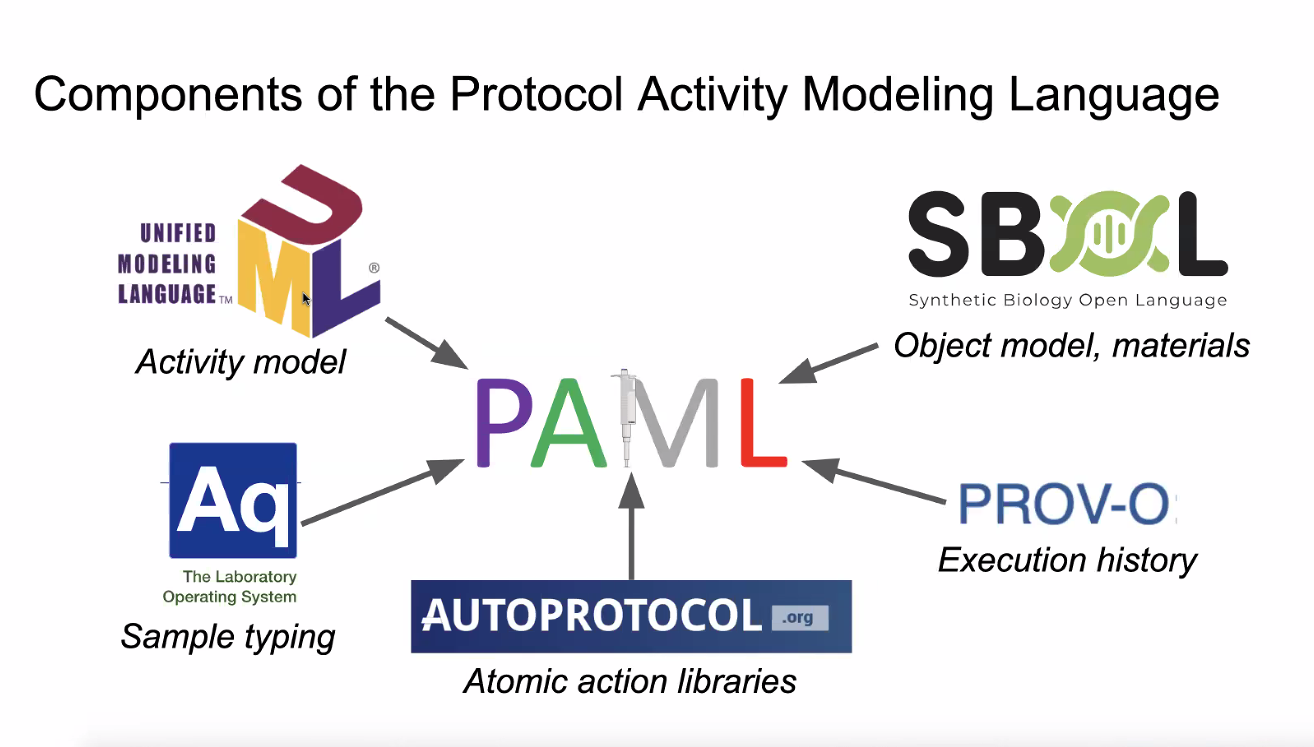
### Agenda
1. Define our challenge problem more concretely.
1. ~~Set high-level OKRs and milestones.~~
1. Coordinate further comms and project management.
### Notes
- Objectives
- Define our challenge problem more concretely.
- Scope: Known-good reproducibility demo of a Golden Gate protocol
- Options:
- Marburg with BsmBI
- https://www.protocols.io/view/golden-gate-lvl-0-8edhta6
- Marburg with BsaI-HFv2
- https://drive.google.com/file/d/1TIfXaeupHi2ZGUXDSk8RGXE9tC3q9e_w/view?usp=sharing
- Intended Goal: Marburg with BsaI, forked from the Marburg with BsmBI protocol:
- Our fork is here
- https://www.protocols.io/view/golden-gate-lvl-0-b2k4qcyw
- Coordinate further comms and project management.
- Friendzymes has github and discord-oriented comms
- We need to get added to the bioprotocols github org as well
- Jeremy suggests we work where we're admins / it's easy (friendzymes) and deposit deliverables over there (bioprotocols)
- ^Ya boy Tim isn't an admin though
- Can add. If scoped to one main repo there shouldn't be too much spam / problems. <- Ok, sure (https://github.com/tsdobbs)
- Google Group exists but nobody's in it
- https://groups.google.com/g/bioprotocols
- Set high-level OKRs and milestones.
- Bump to Meeting 2
- Medium-term goal for BioProtocols: Get an alpha out by April 2022
- "LIMS-less interlab coordination"
- Links
- PAML basics
- https://github.com/bioprotocols/paml
- https://github.com/bioprotocols/pamled
- https://github.com/bioprotocols/paml-specification
- https://github.com/rpgoldman/container-ontology
- [Data Representation in the DARPA SD2 Program](https://doi.org/10.1101/2021.09.17.460644)
- [Process-Level Representation of Scientific Protocols with Interactive Annotation](https://arxiv.org/abs/2101.10244)
- [2021-10-14_COMBINE2021_Standards-Meeting.pdf](https://files.catbox.moe/pvr6wb.pdf)
- This working group
- [friendzymes/community #14 - Meeting: PAML Pilot](https://github.com/friendzymes/community/issues/14)
- [friendzymes project board - PAML](https://github.com/orgs/friendzymes/projects/5/views/8)
### Action Items
- [ ] isaacG update forked protocol
- [ ] forked protocol to PAML (github)
- [ ] research and summary updates on other enzymes
- [ ] Scott to talk to Jenny / Nico to build a list of tested combinations to express GFP
- [ ] isaacG whenisgood for next meet, prior to christmas and preferably 2021-12-11 - 2021-12-18