# Technical Monitoring (notes)
:::info
- **Date:** January 21, 2025 @ 12 pm EST
- **Participants:** OE, CM, MH, EM, MG, JHL, TS
:::
:::success
## Agenda
1. *NiPreps Hack* (NSC reporting)
* Dates, location, overall goals/projects
* Formally invite all TechMon attendees
* Trigger brainstorming
* https://hackmd.io/I68ejqi0Tf61W88K0RUyZw
1. fMRIPrep telemetry with Sentry
* Analysis code: https://github.com/nipreps/fmriprep_stats/blob/master/notebooks/fmriprep_stats.ipynb
* Success rate slowly receding
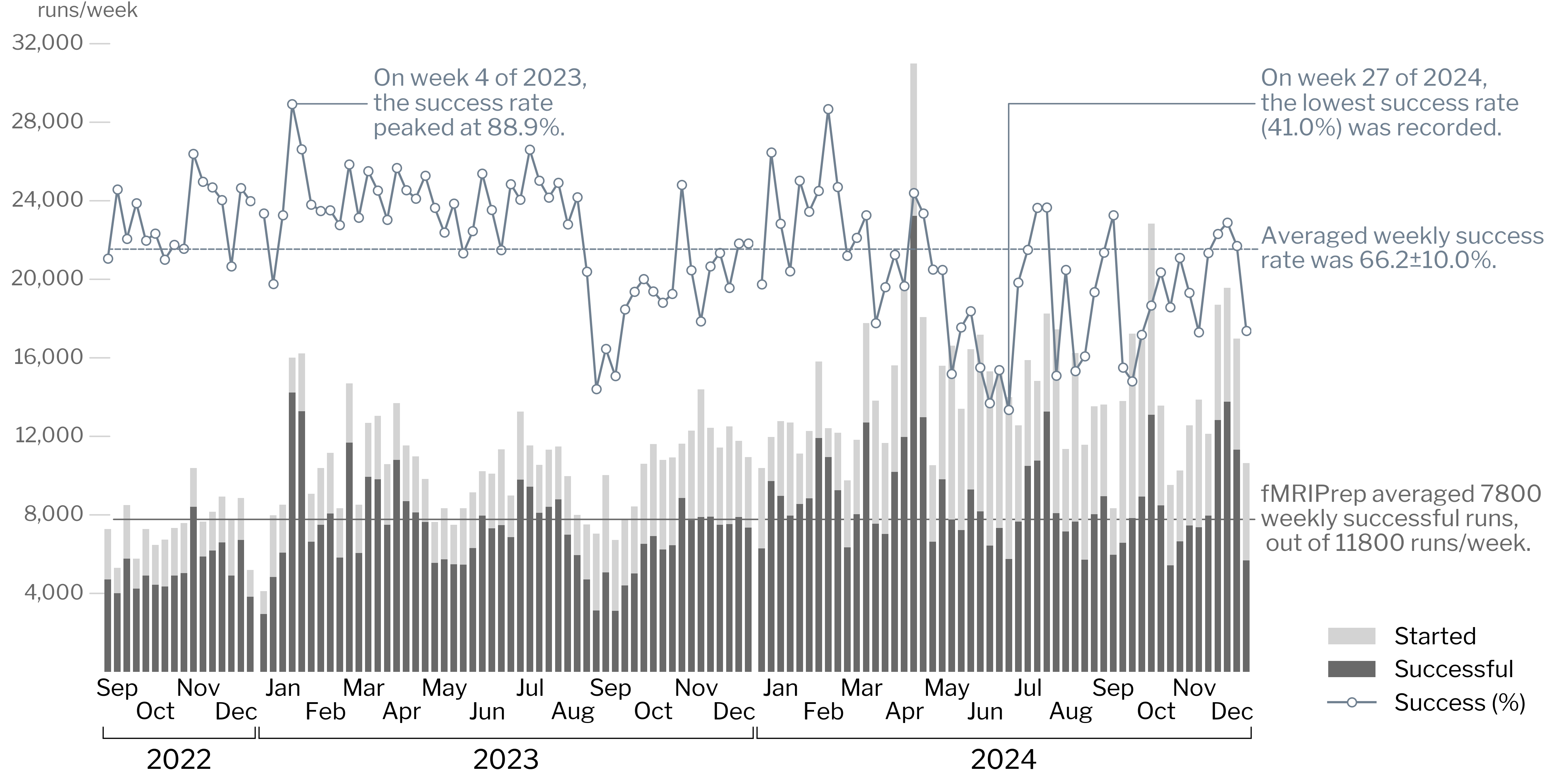
* Starting with 23.x versions
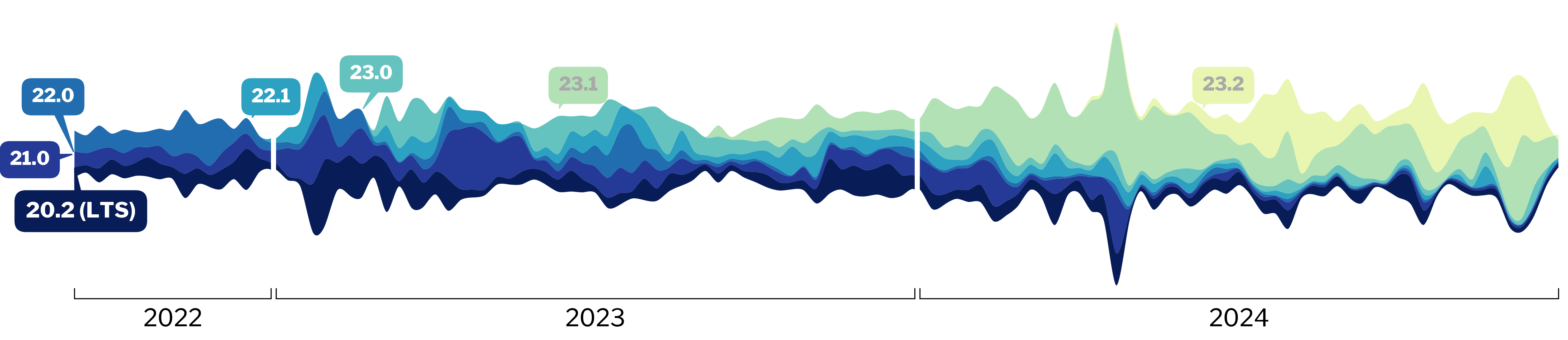
* [nipreps/fmriprep#3419](https://github.com/nipreps/fmriprep/issues/3419)
(We do not register 24.x success pings)
* Change in Sentry API for submission?
* Change in Sentry API for retrieval?
* Regression?
* Failure?
* Same trend with Migas?
* Where are the error reports being centralized? [Question from the ignorance JHL]
1. Migration to Migas
* Are we ready?
* If not, can we automate Sentry analytics?
1. *NiFreeze*
* [NEW] Refactor of the dataset data structure [nipreps/nifreeze#52](https://github.com/nipreps/nifreeze/pull/52).
* [NEW] [nipreps/nifreeze#62](https://github.com/nipreps/nifreeze/pull/62) [Proposal of new model to stack estimators](https://github.com/nipreps/nifreeze/issues/12#issuecomment-2598471780):
* The feature/problem: head motion estimation is usually run in several *passes* (levels), where in the beginning very fast and coarse registration settings initialize more precise (and slow) later levels with more sophisticated transforms and generation models.
For example, you'd typically run a very fast head motion estimation with a downsampled version of the dataset and a relatively simple model (e.g., DKI), followed by our GPR and a low-degree polynomial or bspline transform for eddy at the full resolution of the image.
* Currently, we have a loop and several parameters (model, align_kwargs, etc.) must be synchronized. The approach has demonstrated very brittle and hard to maintain.
* A simpler approach of stacking estimators is being proposed:
```Python
estimator_level1 = Estimator(model="b0", ...) # 6-dof
estimator_level2 = Estimator(
model="b0",
input=estimator_level1,
...,
) # 9-dof registration
estimator_level3 = Estimator(
model="gpr",
input=estimator_level2,
...,
) # bspline registration
estimator_level3.fit(dataset_object)
```
This works because the dataset object keeps the status,
and the transforms are updated properly.
* Presubmission inquiry sent to *Nat Comput Sci*.
* Archiving date on *eddymotion* repo needs fixing.
1. Type Hints & pytest error on warning
* [nipreps/nifreeze#24](https://github.com/nipreps/nifreeze/issues/24)
* [nipreps/nifreeze#28](https://github.com/nipreps/nifreeze/pull/28). Any chance to review @effigies?
* Error on warning: [nipreps/nireports#156](https://github.com/nipreps/nireports/pull/156),
[nipreps/nireports#157](https://github.com/nipreps/nireports/pull/157),
[nipreps/nifreeze#40](https://github.com/nipreps/nifreeze/pull/40),
[nipreps/mriqc#1370](https://github.com/nipreps/mriqc/pull/1370). `nifreeze` PR was merged: @effigies mind sparing some time to tell your opinion on this?
1. *Nireports*:
* Resolving a ToDo task: requires review: [nipreps/nireports#151](https://github.com/nipreps/nireports/pull/151) @eilidhmacnicol @oesteban
1. Transition default branch names across *NiPreps* to `main`
* Including any dataset repositories, e.g. [nipreps-data/tests-nifreeze](https://gin.g-node.org/nipreps-data/tests-nifreeze)
1. Some workflows using `actions/cache` not saving cache properly: [nipreps/nireports#161](https://github.com/nipreps/nireports/issues/161), [nipreps/sdcflows#475](https://github.com/nipreps/sdcflows/pull/475)
1. Nibabies XFM outputs
* multi-step registration currently concatenated into single composite transform
* TS: Pre-change, anat-to-MNI disassembled into affine_00, warp_01 and MNI-to-anat disassembled into warp_00, affine_01.
* TS: Post-change, anat-to-MNI disassembles into affine_00, warp_01, affine_02, warp_03 and MNI-to-anat disassembles into affine_00, warp_01, affine_02, warp_03 (not warp first).
* TS: Meeting with MG, Erik Lee, Matt Cieslak later today.
* workaround for nitransforms loading (https://github.com/nipreps/nibabies/pull/430) - should be ported over?
1. McKenzie's white paper (response to reviews deadline: Jan 31).
:::
::: warning
## Topics pushed away to next week
1. MRIQC documentation
* *group* analysis when no level is specified
* *group* analysis on an *individual* analysis
:::
## Notes
### Telemetry
* The [Sentry dashboard](https://docs.sentry.io/product/dashboards/) inspection shows that the events are being gathered, and there is successful executions that are shown with the 24.x version, so the most likely hypothesis is that [the notebook](https://github.com/nipreps/fmriprep_stats/blob/master/notebooks/fmriprep_stats.ipynb) is not working properly.
* There is a private Sentry dahsboard that gathers the collected information and displays it in a friendly way.
* Transition to Migas: we never tested Migas against the Sentry (the latter being considered as a ground truth), so that would need to be checked prior to transitioning.
* MG: prepare some stats for next TechMon.
### NiFreeze
* Have not heard about the presubmission inquiry yet.
* Model for stacking estimators
* Uncertain the best software engineering approach about the data/data representation entities when modifying the intermediate results. Suggestions/reviews are welcome.
* [Ongoing PR](https://github.com/nipreps/nifreeze/pull/62).
### Default branch names
* Have `master` as theirs
* Code: [nipreps/dmriprep](https://github.com/nipreps/dmriprep), [nipreps/fmriprep](https://github.com/nipreps/fmriprep), [nipreps/fmriprep-rodents](https://github.com/nipreps/fmriprep-rodents), [nipreps/micro-nipype](https://github.com/nipreps/micro-nipype), [nipreps/migas-server](https://github.com/nipreps/migas-server), [nipreps/mriqc](https://github.com/nipreps/mriqc), [nipreps/mriqcwebapi](https://github.com/nipreps/mriqcwebapi), [nipreps/nibabies](https://github.com/nipreps/nibabies), [nipreps/nirodents](https://github.com/nipreps/nirodents), [nipreps/niworkflows](https://github.com/nipreps/niworkflows), [nipreps/nondefaced-detector](https://github.com/nipreps/nondefaced-detector), [nipreps/sdcflows](https://github.com/nipreps/sdcflows), [nipreps/smriprep](https://github.com/nipreps/smriprep)
* Data: [nipreps-data/bcp](https://gin.g-node.org/nipreps-data/bcp), [nipreps-data/bcp-derivatives](https://gin.g-node.org/nipreps-data/bcp-derivatives), [nipreps-data/bootcamp-geneva-2024](https://gin.g-node.org/nipreps-data/bootcamp-geneva-2024), [nipreps-data/brain-extraction-tests](https://gin.g-node.org/nipreps-data/brain-extraction-tests), [nipreps-data/hcph-mini](https://gin.g-node.org/nipreps-data/hcph-mini). [nipreps-data/hbn-mriqc](https://gin.g-node.org/nipreps-data/hbn-mriqc), [nipreps-data/HCP101006](https://gin.g-node.org/nipreps-data/HCP101006), [nipreps-data/HCP101006-derivatives](https://gin.g-node.org/nipreps-data/HCP101006-derivatives), [nipreps-data/tests-nifreeze](https://gin.g-node.org/nipreps-data/tests-nifreeze)
* CM: Many GHA workflow files have the `master` name hard-coded as the target branches, so maybe prior to renaming both `main` and `master` should be added.
 Sign in with Wallet
Sign in with Wallet

