<center><h1>Slides - 1</h1></center>
1. **Differentiate between DNA metabolic processes ( replication , transcription , translation ) , and identify their intracellular locations**
<img src="https://39363.org/IMAGE_BUCKET/1663046808756-879611715.png" style="zoom:50%;" />
2. **Understand the structures and chemical differences between DNA and RNA**
- DNA is "de-oxy" nucleic acid
- Doesn't have a $-OH$ group on carbon $2^{'}$
- RNA is "ribo" nucleic acid
- Has a $-OH$ group on carbon $2^{'}$
<img src="https://39363.org/IMAGE_BUCKET/1663047944920-500052824.jpeg" style="zoom:50%;" />
3. **Classify nitrogenous bases as purines or pyrimidines**
- Purines = Guanine , Adenine
- "Pure as gold ( A U )"
- Pyrimidines = Thymine , Cytosine
- "C U T the pie"
<img src="https://39363.org/IMAGE_BUCKET/1663048843959-364094058.png" style="zoom:50%;" />
4. **Identify and distinguish between phosphorylated forms of the nitrogenous bases**
- mono , di , tri
5. **Explain the molecular basis of tautomerization**
- Interconversion between two structural isomers of a molecule
- The two isomers are called tautomers
<img src="https://39363.org/IMAGE_BUCKET/1663047832214-295647891.jpeg" style="zoom:33%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663049416008-83914372.jpeg" style="zoom:67%;" />
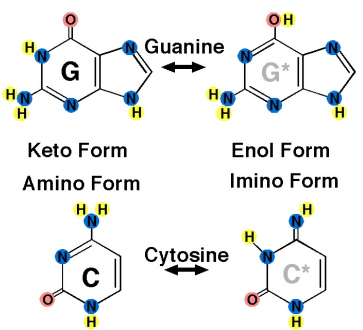
6. **Know structures , names , and symbols ( both two letter and four letter ) of Deoxyribonucleotide and Ribonucleotide nomenclature**
<img src="https://39363.org/IMAGE_BUCKET/1663048046851-529415592.jpeg" style="zoom:50%;" />
7. **What is it about the structure of nucleotides that enables them to absorb UV light?**
- Nucleotides have rings that are : aromatic / conjugated $\pi$-electrons
- Ultraviolet ( UV ) light is electromagnetic radiation that corresponds to wavelengths between 200 nm and 400 nm.
- UV absorption causes an electron transition from the ground state to a higher energy level.
- In the ground state :
- $\pi$ electrons from double bonds are in the $\pi$ bonding molecular orbital
- nonbonding electrons are in the $n$ nonbonding molecular orbital.
- These electrons can be excited to the $\pi^*$ antibonding orbital
<center><img src="https://39363.org/IMAGE_BUCKET/1645899268273-688640030.png" style="zoom:100%;"/>6</center>
<center><img src="https://39363.org/IMAGE_BUCKET/1645899268273-688640030.png" style="zoom:100%;"/></center>
- The lower the energy gap between HOMO and LUMO , the easier it is to excite electrons
- energy gap $\frac{1}{\propto}$ wavelength
- Electrons have more "space" in a conjugated aromatic ring
- Less likely to break bonds when excited
<img src="https://39363.org/IMAGE_BUCKET/1663048379366-119866904.png" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663048429489-394623114.png" style="zoom:67%;" />
- https://socratic.org/questions/why-do-aromatic-rings-absorb-light
- https://chem.libretexts.org/Bookshelves/Organic_Chemistry/Map%3A_Organic_Chemistry_(Smith)/14%3A_Conjugation_Resonance_and_Dienes/14.15%3A_Conjugated_Dienes_and_Ultraviolet_Light
- https://www.quora.com/Why-do-aromatic-side-chains-absorb-UV-light-Why-is-tryptophan-the-most-absorptive-of-UV-light/answer/Cameron-Morrison?ch=10&oid=73315423&share=c87404b4&target_type=answer
<center><img src="https://39363.org/IMAGE_BUCKET/1663095668515-863519929.jpeg" style="zoom:100%;" /></center>
<center><img src="https://39363.org/IMAGE_BUCKET/1663095698719-532590942.jpeg" style="zoom:80%;" /></center>
<center><img src="https://39363.org/IMAGE_BUCKET/1663093116835-627705802.jpeg" style="zoom:30%;" /></center>
<center><img src="https://39363.org/IMAGE_BUCKET/1663093165770-834891020.jpeg" style="zoom:40%;" /></center>
8. **If there is 36% guanine , then percent of rest of bases = ?**
- C = 36%
- A = 14%
- T = 14%
9. **Problem : Some proteins bind to ( “read” ) specific DNA or RNA nucleotide sequences , while other proteins bind randomly to any DNA or RNA sequence. What types of interactions would you expect to be involved in each type of binding?**
- Both sequence-specific and random binding occur by hydrogen bonding
10. **What types of inhibition might be at work in figure 22-37?**
<img src="https://39363.org/IMAGE_BUCKET/1663053317775-478533001.png" style="zoom:50%;" />
- AMP , and GMP inhibit glutamine-PRPP amidotransferase in concerted inhibition
- IMP inhibits glutamine-PRPP amidotransferase via sequential feedback inhibition
- Excess GMP inhibits formation of xanthylate from inosinate by IMP dehydrogenase
- Excess AMP inhibits formation of adenylosuccinate by adenylosuccinate synthetase and ribobse pyrophosphokinase
**What are the consequences of these regulatory loops for individual nucleotide levels?**
- Balances nucleotide levels via allosteric regulation
11. **What sources provide substrates for nucleotide salvage?**
- Uracil is slavaged to uridine
- Thymine is salvaged to thymidine
- Cytidine is salvaged from uracil pathway
- Adenosine is converted to inosine
- IMP can be converted into GMP
12. **Summary / Highlights of Purine Nucleotide Synthesis**
- In contrast to pyrimidine synthesis , the heterocyclic ring is built on the "activated sugar" ribose phosphate foundation
- A critical intermediate linking de novo synthesis to salvage synthesis is PRPP
- Amino acids used in purine synthesis are glycine , aspartate , and glutamine
- One-carbon units are donated by tetrahydrofolate cofactors
- In de novo synthesis , IMP is synthesized first , then converted to AMP and GMP
- The salvage of purine bases occurs via :
- PRPP and HGPRTase / APRTase
- Nucleoside kinases
- Salvage via PRPP
- important quantitatively
- most PRPP is used in salvage pathway
- Deficiency of HGPRTase
- nitrigenous bases will be extrected not salvage
- de novo synthesis is favored
- leads to uric acid build up and gout
- PRPP concentration regulates the entire de novo pathway
13. **Summary / Structures and Enzymes to Remember in Purine Nucleotide Synthesis**
- Purines :
- PRPP
- IMP
- AMP , ADP , ATP ( and deoxy forms )
- GMP , GDP , GTP ( and deoxy forms )
- Pyrimidines :
- CMP ( and deoxy forms )
- TMP ( and deoxy forms )
- UMP
- Enzymes:
- PRPP synthetase
- HGPRTasee , APRTase , OPRTase
- dihydrofolate reductase
- ribonucleotide reductase
- thymidylate synthase
- xanthine oxidase
14. **Problem : One form of severe combined immunodeficiency ( SCID ) disease is an autosomal recessive disease of childhood in which extremely high levels of deoxyadenosine are toxic to the formation of immune system B cells and T cells.**
**Which of the purine salvage enzymes is defective in this form of the disease?**
- Adenosine Deaminase ( ADA )
**Why is this disease recessive?**
- autosomal recessive pattern = both copies of a particular gene contain defects
- one inherited from the mother and one from the father
- Females have two X-chromosomes
- The mother will not be affected by carrying only one abnormal X-chromosome ,
- Her male children will have a 50% chance of being affected with the disorder by inheriting the faulty gene.
- Her female children will have a 50% chance of being carriers for the immunodeficiency
- https://www.niaid.nih.gov/diseases-conditions/severe-combined-immunodeficiency-scid
<img src="https://39363.org/IMAGE_BUCKET/1663057566771-228513935.jpeg" style="zoom:24%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663057483417-43182729.jpeg" style="zoom:25%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663057546500-839841982.jpeg" style="zoom:24%;" />
15. **Summarize the similarities and differences between purine and pyrimidine de novo synthesis in terms of the strategy for assembly of the heterocyclic rings , amino acid requirements , role of PRPP , and folate co-factors**
- Pyrimidine :
- the pyrimidine ring is built <u>before</u> the ribose phosphate is added
- OPRTase is similar to HGPRTase and APRTase
- PRPP is used to convert orate into Orotidine monophsphate ( OMP )
- Tetrahydrofolate is oxidized to dihydrofolate during TMP synthesis , and reduced by DHFR
- Amino Acids Required = glutamine and aspartate
- aspartate adds to carbamoyl phosphate
- Salvage is primarily via nucleoside kinases or "reversal" of nucleoside phosphorylase reactions
- https://microbenotes.com/de-novo-pyrimidine-synthesis/
- Purine :
- PRPP catalyzes the first committing step in de novo purine synthesis
- PRPP is the base substrate
- https://microbenotes.com/purine-synthesis/
- Amino Acids Required = glycine , aspartate , glutamine
- PRPP is synthesized from ribose-5-phosphate in the pentose phosphate pathway
- PRPP synthetase is the rate limiting step in de novo synthesis of purines and pyrimidines
16. **Contrast the roles of PRPP in de novo nucleotide synthesis vs salvage synthesis**
- $[\ \text{HGPRTase}\ ]\ \frac{1}{\propto}\ [\ \text{PRPP}\ ]$
- $[\ \text{PRPP}\ ] \propto \text{Purine Production}$
- If $[\ \text{HGPRTase}\ ]$ == High :
- $[\ \text{PRPP}\ ]$ Decreases
- Purine synthesis decreases
- Purine salvage increases
- If $[\ \text{HGPRTase}\ ]$ == Low :
- $[\ \text{PRPP}\ ]$ Increases
- Purine synthesis increases
- overproduction of purines will broken down and excreted as uric acid
- excess $[\ \text{Uric Acid}\ ]$ ➡️ gout
- Purine salvage decreases
- Purines are synthesized starting with PRPP being converted to phosphoribosylamine
17. **Recognize the numbering of nitrogenous bases and sugars in DNA and RNA**
<img src="https://39363.org/IMAGE_BUCKET/1663208354327-81974903.jpeg" style="zoom:50%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663208294065-840680710.jpeg" style="zoom:50%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663208437713-242332670.png" style="zoom:67%;" />
18. **Explain the use of nucleotide antimetabolites in cancer chemotherapy**
- Antimetabolites interfere with DNA production , therefore cancerous cells cannot divide
- Alters nucleotides or deletes supply of deoxynucleotides.
- Methotrexate blocks production of folate. Folate is needed for Glycine production
- https://en.wikipedia.org/wiki/Antimetabolite#Cancer_treatment
19. **Explain the use of folate analogs in cancer chemotherapy**
- Methotrexate , Trimethoprim , and other anti-folates inhibit the production of active tetrahydrofolate ( THF ) from the inactive dihydrofolate ( DHF )
- this interferes with folate metabolism
- cells that divide rapidly need a high level of folate
- cell division requires DNA synthesis
- folate is used to make purines and pyrimidines
- https://en.wikipedia.org/wiki/Folate#DNA_synthesis
<img src="https://39363.org/IMAGE_BUCKET/1663211171216-176113477.png" style="zoom:67%;" />
20. **Identify the common rationale underlying the biochemical feedback mechanisms of IMP anabolism , purine biosynthesis de novo , and Ribonucleotide Reductase**
- IMP Anabolism :
- allosteric regulation
- De Novo Purine Biosynthesis :
- PRPP amidotransferase is inhibitied by AMP , GMP , IMP
- https://step1.medbullets.com/biochemistry/102004/de-novo-nucleotide-synthesis
- Ribonucleotide Reductase :
- Depending on the allosteric configuration, one of the four ribonucleotides binds to the active site
- Regulation of RNR is designed to maintain balanced quantities of dNTPs
- When ATP binds to the allosteric activity site , it activates RNR
- In contrast , when dATP binds to this site, it deactivates RNR
- The allosteric mechanism also regulates the substrate specificity and ensures the enzyme produces an equal amount of each dNTP for DNA synthesis
- https://en.wikipedia.org/wiki/Ribonucleotide_reductase#Regulation
<img src="https://39363.org/IMAGE_BUCKET/1663212731430-614520522.jpeg" style="zoom:67%;" />
21. **Understand the biochemical reasons for the clinical manifestations of HGPRTase deficiency**
- Kelley-Seegmiller Syndrome : deficiency causes high levels of uric acid in the blood. leads to uric acid stones in urinary tract
- Lesch-Nyhan Syndrome : HPRT1 mutation on X chromosome.
- Build up of uric acid in all body fluids.
- Causes gout and kidney problems.
- Poor muscle control and intellectual disability.
- Self mutilation
22. **Problem : Why are some drugs that are used for cancer chemotherapy also useful against certain autoimmune diseases?**
- Methotrexate is an antimetabolite and antifolate chemotherapy drug that directly kills cells by inhibiting the metabolism of folate , an essential B vitamin used to synthesize DNA
- Rheumatoid arthritis (RA) can be treated with methotrexate
- https://www.ons.org/pubs/article/224956/download
23. **Effect of Fluor-dUMP on thymidylate synthase**
- FdUMP = suicide inhibitor of thymidylate synthase
- Fluorine refuses to leave once bound
- Cellular Uptake of FdUMP
- $3^{'} \ce{->} 5^{'}$ exonucleolytic activity to release $N$ FdUMP molecules from each molecule of FdUMP[ $N$ ]
- Inhibition of thymidylate synthase ( TS ) by FdUMP
- when TS is inhibited , there is a decrease of dTMP
- because of this , dTTP ( the precursor nucleotide of DNA ) will not be synthesized either
- https://en.wikipedia.org/wiki/Fluorodeoxyuridylate#Consequences_of_the_TS_inhibition
24. **Why does FdUMP kill cancer cells?**
- It irreversibly binds and inactivates thymidylate synthase
- Blocks synthesis of dTTP
- If you can't make DNA , you can't undergo mitosis.
- https://moscow.sci-hub.se/2788/be246e2358091fa29551c707f9181cfc/gmeiner1999.pdf#navpanes=0&view=FitH
25. **Highlights of Pyrimidine Nucleotide Synthesis**
- The pyrimidine ring is built before the ribose phosphate is added
- Amino acids used in pyrimidine synthesis are aspartate and glutamine
- OPRTase is similar to HGPRTase and APRTase
- Tetrahydrofolate is oxidized to dihydrofolate during TMP synthesis , and reduced by DHFR
- UMP is converted to CMP or TMP
- Salvage is primarily via nucleoside kinases by "reversal" of nucleoside phosphorylase reactions
26. **Deoxyribonucleotdies are synthesized ONLY by reduction of ribonucleoside diphosphates**
27. **How can the same nucleotide have two different regulatory effects?**
- Depending on the allosteric configuration , one of the four ribonucleotides binds to the active site
- https://en.wikipedia.org/wiki/Ribonucleotide_reductase#Regulation
28. **Why might these concentrations be so different from one another ?**
- ATP = $2 - 10\ \text{mM}$
- rNTPs = $0.05 - 2\ \text{mM}$
- dNTPs = $2 - 60\ \mu M$
- $mM = 1 * 10^{-3}$
- $\mu M = 1 * 10^{-6}$
- $\therefore\ :\ [\ \text{dNTPs}\ ] < [\ \text{rNTPs}\ ]$
- dNTPs are lower in concentration than rNTPs because they are constantly being used up in DNA replication.
29. **Why is fluorouracil called a "suicide inhibitor" ?**
- It irreversibly binds and inactivates thymidylate synthase
30. **Hydroxyurea acts as a free radical scavenger that arrests human cells in the DNA synthetic phase ( S phase ) of the cell cycle and inhibits the repair of damaged DNA. Why is ribonucleotide reductase an important target of this drug ?**
- Hydroxycarbamide inhibits ribonucleotide reductase by scavenging tyrosyl free radicals
- This decreases the production of deoxyribonucleotides
- Tyrosyl free radicals are involved in the reduction of nucleoside diphosphates ( NDPs )
- https://en.wikipedia.org/wiki/Hydroxycarbamide#Mechanism_of_action
- Hydroxyurea selectively inhibits ribonucleoside diphosphate reductase
- Ribonucleoside diphosphate reductase = an enzyme required to convert ribonucleoside diphosphates into deoxyribonucleoside diphosphates,
- This prevents cells from leaving the G1/S phase of the cell cycle.
- If you can't make DNA , you can't leave the cell cycle
- Hydroxyurea also exhibits radiosensitizing activity by maintaining cells in the radiation-sensitive G1 phase and interfering with DNA repair.
- https://en.wikipedia.org/wiki/Hydroxycarbamide#Pharmacology
31. **Problem : At the level of nucleotide synthesis , why does hydroxyurea arrest human cells in the DNA synthetic phase ( S phase ) of the cell cycle and inhibit the repair of damaged DNA ?**
- Causes DNA double strand breaks near replication forks
- Its also a Nucleic Acid Synthesis Inhibitor
32. **Interpret the experiments implicating DNA in inheritance**
- Griffith :
- Something had passed from heat killed bacteria to the nonvirulent $R$ strain , making them virulent
- he called it the "transforming principal"
- Hershey and Chase :
- Bacteria phages grown with <u>protein coats</u> with radiolabeled sulfur did NOT HAVE radioactive sulfur found inside the cell.
- Bacteria phages with their <u>DNA radiolabeled</u> HAD radioactive sulfur found in the cell.
- Proves proteins are NOT genetic material
- Proves DNA IS genetic material
- Avery , MacLeod and McCarty :
- A small amount of heat killed "S" type pneumococcus was able to be conjugated into "R" type pneumococcus and cause the transformation from $R$ to $S$ serotypes
- Proved it wasn't a protein
- there would be way more phosphorous
- Proved it was DNA by tests with DNAase
- when DNAase was present , it chewed up the S-DNA and therefore only $R$ strain was found , not the $S$ strain
<img src="https://39363.org/IMAGE_BUCKET/1663219924888-11546613.jpeg" style="zoom:67%;" />
33. **Summarize the structure of DNA and the forces that stabilize that structure**
- B form DNA is a right-handed doubled stranded helix of deoxyribonucleotides
- It will have Negative Supercoiling
- Stabilization Forces :
- Hydrogen Bonds
- $\pi - \pi$ base stacking
- Sodium cations stabilize negative charged DNA
34. **Attribute a mechanism of inheritance based on the structure of DNA**
- DNA can be supercompacted into nucleosomes ➡️ chromatin ➡️ chromosome
- chromosomes cross over during meiosis
- In eukaryotes , reproductive cells ( gametes ) are produced via meiosis , during which a parent cell ( 2n ) divides to produce four genetically distinct daughter cells containing half the original number of chromosomes ( n ).
- Meiosis has four phases ( like mitosis ) , but these phases occur in two successive cell division stages ( meiosis I and II ).
- In prophase I of meiosis , homologous chromosomes recognize each other and line up side by side during synapsis.
- Because each homologous chromosomes consists of two identical ( sister ) chromatids , this adjacent chromosomal alignment forms a tetrad ( four chromatids total ).
- Tetrads arise when a synaptonemal complex ( protein structure ) forms between homologous chromosomes and holds them together tightly.
- The tetrad structure allows physical contact between the paternal and maternal chromosomes at the chiasma , a point where a chromosome segment can break off and rejoin with the opposite homologous chromosome.
- This exchange of DNA is the hallmark feature of the process called crossing over.
- Crossover events result in daughter cells with chromosomes containing combinations of alleles that differ from those in the parent cell , leading to eukaryotic genetic recombination and increased genetic diversity.
- Because crossover events produce genetically unique gametes , the offspring that develops after fertilization is genetically different from either parent and more genetically diverse.
<img src="https://39363.org/IMAGE_BUCKET/1641588395522-661270689.png" style="zoom:67%;"/>
<img src="https://39363.org/IMAGE_BUCKET/1641588666778-765098040.png" style="zoom:67%;"/>
<img src="https://39363.org/IMAGE_BUCKET/1663220871079-941919841.jpeg" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663220937767-121465956.jpeg" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1626983849119-235266385.png" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663221087439-851245600.jpeg" style="zoom:67%;" />
35. **Compare the structures of DNA 'isomers'**
- B Form :
- Major Groove = Wide and Deep
- Minor Groove = Narrow and Less Deep
- Super Coiling = Negative
- Right Handed
- A Form :
- Major Groove = Deeper
- Minor Groove = Shallower and Broader
- Super Coiling = Negative
- Right Handed
- C Form :
- Major Groove = Deeper
- Minor Groove = Shallower and Broader
- Super Coiling = Negative
- Right Handed
- Z Form :
- Major Groove = Flat / Non-Existant
- Minor Groove = Very Narrow and Deep
- Super Coiling = Positive
- Left Handed
36. **Explain how specific nucleotide sequences can affect DNA structure**
- Palindromic DNA or RNA can cause hairpins and cruciforms
37. **How do we know that DNA is the material of inheritance ?**
- Griffith heat-killed virulent bacteria conjugated to live encapsulated virulent bacteria mouse experiments
38. **Problem : A researcher introduces jellyfish DNA that contain the genes for a cyan ( blue ) fluorescent protein or a yellow fluorescent protein into human cells , with results shown below.**
<img src="https://39363.org/IMAGE_BUCKET/1663030595964-100129401.png" style="zoom:50%;" />
**In what way( s ) do these results resemble the results of Griffith , and those of Avery , MacLeod and McCarty?**
- GFP can be included in a plasmid expressing other genes to indicate a successful transfection of a gene of interest
- Similar to Avery et al. study where foreign DNA was incorporated.
- In this example : transfection of a gene changed the cell's phenotype
- In the Griffith example : heat killed virulent DNA was transfected and made the cell change its phenotype
- In the Avery et al. Paper : DNA was transfected and made pneumococcus change serotypes from ( R ) to ( S )
- https://en.wikipedia.org/wiki/Green_fluorescent_protein#Fluorescence_microscopy
39. **The density of DNA can be measured by centrifugation , and is dependent on nucleotide composition**
- GC Base pair stacking is more stable than AT base pair stacking
- this can be used to separate DNA
- Put a heavy salt ( CsCl ) in a tube , with DNA
- the CsCl can sedimate toward the bottom of the tube
- the density the DNA follows is toward the bottom
- the DNA travels toward the interface of CsCl and solvent solution
- low GC content forms a band toward the top of the tube
- high GC content forms band toward the interface
40. **The stability of DNA against unwinding of the double helix is dependent on nucleotide composition**
- $G-C$ pairs have 3 hydrogen bonds and are more stable than $A-T$ pairs with 2 hydrogen bonds
41. **Stacking interactions play a dominant role in determining DNA stability**
- GC Base pair stacking is more stable than AT base pair stacking
- vertical $\pi - \pi$ / induced dipole stacking
42. **$T_m$ = temperature at the midpoint of the melting transition**
- when 50% of the bases have unstacked
- roughly proportional to the $G + C$ content
$$
T_m = \left(\ \left( \left( 81.5 + \left(\ 16.6 * log\left(\ \frac{[Na^+]}{1.07 * [Na^+]} \right) \ \right) \right) + \left(\ 0.41 * \left(\ \%G + \%C \ \right) \ \right) \ \right) - \left(\ \frac{500}{\text{length}} \ \right)\ - \left(\ \text{\% mismatch}\ \right)\right)
$$
43. **Predict the effect on $T_m$ :**
- Increasing $\text{ [ salt ]}$ :
- Raises the melting temperature
- Stabilizes DNA
- Increasing $\left(\ G + C \ \right)\ \text{base pairs}$ :
- $GC$ rich melts later / at higher temperatures than $AT$
- Increasing DNA length :
- as the length of DNA increases , then melting temperature increases
- if you have a very long DNA molecule , the chances of reannealing are higher
- Increasing % mismatched base pairs :
- melting temp is inversely proportional of percent of mismatches
- if you have 3 DNA molecules , 2 are exactly complementary , and 1 is 98% complementary
- the DNA with perfect complementary has a higher melting temperature than the imperfect one
44. **Problem :**
- The polymerase chain reaction ( PCR ) requires that short single strands of DNA ( oligonucleotides ) anneal specifically to their complementary sequence in a denatured mixture of the total genomic DNA sequences of an organism.
- What conditions would favor specific annealing of the oli
- Which conditions would favor specific annealing of the oligonucleotides to their EXACT complementary sequences and NOT to partially mismatching sequences?
- A = Low salt , & annealing temperature below $T_m$
- B = High salt , & annealing temperature below $T_m$
- <span style="border: 2px solid green;">C = Low salt , & annealing temperature near $T_m$</span>
- D = High salt , & annealing temperature near $T_m$
- E = Low salt , & annealing temperature above $T_m$
- F = High salt , & annealing temperature above $T_m$
- high salt stabilizes DNA and makes it easy to base pairs
- lower salt destabilizes DNA
- annealing temperature near $T_m$ makes it difficult for strands to anneal
- perfectly complementary strands are more likely to anneal at this temperature than imperfectly base paired strands
- you want to make salt conditions and annealing temp so difficult that you can make the tiny distinction between imperfect base pairing and perfect base pairing
<center><h1>Slides - 2</h1></center>
1. **Explain the relationship between copy number of a DNA sequence family and its annealing rate**
- Copy Number = number of copies of a particular DNA sequence in a cell
- $\text{Copy Number} \propto \text{Annealing Rate}$
- More copies = higher probability of base pairing
- if primers attach to the template strand correctly ➡️ polymerization of DNA ➡️ copy number of the DNA sequence increases
2. **Classify a DNA molecule as a relaxed or supercoiled form**
- Positive Supercoiling :
- supercoils in the left-handed direction
- caused by overwinding $\left(\ >\frac{1\ \text{Turn}}{10\ \text{Base Pairs}} \ \right)$
- Negative Supercoiling :
- supercoils in the right-handed direction
- caused by underwinding $\left(\ >\frac{10\ \text{Base Pairs}}{1\ \text{Turn}} \ \right)$
3. **Predict the effect of DNA supercoiling on the formation of non-B ( non-Watson-Crick ) DNA**
- A and C Form DNA can be induced by negative supercoiling
4. **Infer the structure of restriction enzyme molecules based on their DNA recognition sequences**
- BamH1 :
- Recognition Sequence :
- 5' GGATCC
- 3' CCTAGG
- Cut :
- 5' ---G GATCC--- 3'
- 3' ---CCTAG G--- 5'
- EcoR1 :
- Recognition Sequence :
- 5' GAATTC
- 3' CTTAAG
- Cut :
- 5' ---G AATTC--- 3'
- 3' ---CTTAA G--- 5'
- Hind3 :
- Recognition Sequence :
- 5' AAGCTT
- 3' TTCGAA
- Cut :
- 5' ---A AGCTT--- 3'
- 3' ---TTCGA A--- 5'
- https://en.wikipedia.org/wiki/List_of_restriction_enzyme_cutting_sites
5. **Why is $T_m$ dependent on ionic strength ?**
- melting temp of DNA is proportional to sodium ion concentration
- positive sodium stabilizes negative DNA
- increases melting temperature
6. **What would be the effect of adding an excess of a single sequence of DNA on the annealing rate?**
- $\text{Unique + ssDNA} \ce{<=>} \text{dsDNA:unique + dsDNA}$
- pushes equilibrium to the right
- towards dsDNA
7. **Why does the molecular structure of an electrophoretic gel allow supercoiled DNA to run faster than relaxed DNA?**
- Circular DNA has a harder time moving through than supercoiled DNA
- Relaxed / no topological strain will move most slowly
**Is this consistent with the separation of proteins by MW on SDS-PAGE ?**
- Heaver = more MW = don't migrate as far
- Lighter = less MW = migrate farther
- SDS-PAGE acts via size
- Electrophoresis acts via charge
8. $L_k = T + W$
- $L_k$ = number of times one strand winds around the other
- $T$ = the twist ( Watson-Crick turns ) of the DNA
- $W$ = number of supercoils
9. **Recognize a recombinant DNA molecule**
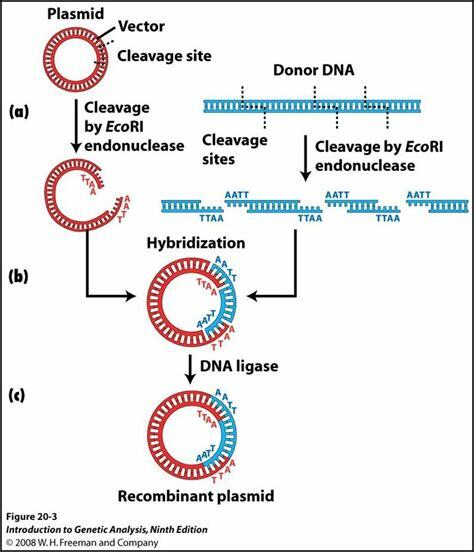
10. **Explain the function of restriction enzymes in forming and characterizing a recombinant DNA molecule**
- Recombinant DNA Molecule = result of in vitro joining of two DNA fragments that would not normally reside together
- since we are using a restriction enzyme , we can be sure that after the cut , both strands of DNA will have the same ends
- the broken apart plasma has sticky ends
- you put in a ligase enzyme , and create a recombinant DNA
- then you put the modified plasmid back into the host cell , and the host cell will make millions of copies of your plasmid
- commercial plasmids have known restriction sites
- it can be done with chromosomal DNA
- the collection of clones = genomic library
11. **Explain the utility of antibiotic resistance in bacterial cloning and how this builds on the work of Avery et al.**
- When creating a plasmid , you can insert an antibiotic resistance cassette
- If you add antibiotic , only cells that have taken up your plasmid will survive ,
- allowing you to "select" for your cell with the plasmid
12. **Summarize the differences between a genomic DNA library and a cDNA library**
- genomic library = collection of clones
- representation for all the genes
- cDNA library = representation for the genes that are expressed in the cell at any given point in time
- synthetic DNA whose base sequences are complementary to genomic DNA
<img src="https://39363.org/IMAGE_BUCKET/1663284122906-6796810.png" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663284164667-236109191.png" style="zoom: 50%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663284062372-499559493.png" style="zoom:67%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663284215404-454018233.jpeg" style="zoom:67%;" />
13. **Explain why the polymerase chain reaction is called a "chain reaction"**
- each cycle adds 1 long product
- but each time you copy and long or short product , you create another short product
- long products increase linearly
- short products increase exponentially
14. **Problem :**
- **DNA structure :**
- **A relaxed ( = no supercoiled ) 3000 bp plasmid DNA molecule is heated to a temperature at which only a 20 bp adenine-thymine region denatures**
- **Using whole integer numbers ( ie not fractions or decimals ) what are the values of $L$ , $T$ , and $W$**
- $L_k = T + W$
- 10 base pairs / Watson crick turn
- A = **before heating ?**
- T = ( 3000 / 10 ) = 300
- W = 0
- L = 300 + 0 = 300
- B = **after heating ?**
- we removed 20 base pairs , aka removed 2 turns
- T = 298
- W = ?
- we haven’t broken the DNA , so L still equals 298
- 300 = 298 + W
- W , therefore = 2
15. **Exonucleases or Endonucleases can cleave circular plasmid DNA?**
- <u>Endo</u>nuclease = cuts internally
- any internal phosphodiesterbonds
- degrades plasmid DNA without ends
- <u>Exo</u>nuclease = cuts from the ends
- degrades linear DNA
16. **What is the difference between a cDNA library and a genomic DNA library ?**
- A cDNA library is made up of complementary DNA , which is DNA that is created from mRNA.
- A genomic DNA library is made up of DNA that is directly taken from an organism's genome.
**Why are these called "libraries" ?**
- The term library is used because the collection of DNA fragments is organized in a way that allows for easy retrieval of the information contained within
- Collection of clones
- library is a collection of DNA fragments that is stored and propagated in a population of micro-organisms through the process of molecular cloning
- https://en.wikipedia.org/wiki/Library_(biology)
- https://en.wikipedia.org/wiki/CDNA_library
17. **How does the transfer of drug resistance relate to the work of Avery , MacLeod , and McCarty ?**
- Avery et al. demonstrated foreign DNA being incorporated and causing genetic transformation
- Plasmids can be created with specific DNA insertions
18. **How can the cloned insert cDNA sequence be recovered ?**
- Use the same restriction enzymes to cut it again
- Sequencing the plasmid that contains the cDNA insert
19. **Identify the three basic steps in a PCR amplification**
- The target DNA is denatured by heating it to 94-96 degrees Celsius. This separates the double stranded DNA into two single strands.
2. Primers (short pieces of complementary DNA) bind to the single strands of DNA at the site of the desired amplification.
3. The temperature is lowered to around 72 degrees Celsius, which is the optimal temperature for DNA polymerase, an enzyme that synthesizes new strands of DNA complementary to the template strand.
<center><img src="https://39363.org/IMAGE_BUCKET/1626984261060-918826333.png" style="zoom:67%;" /></center>
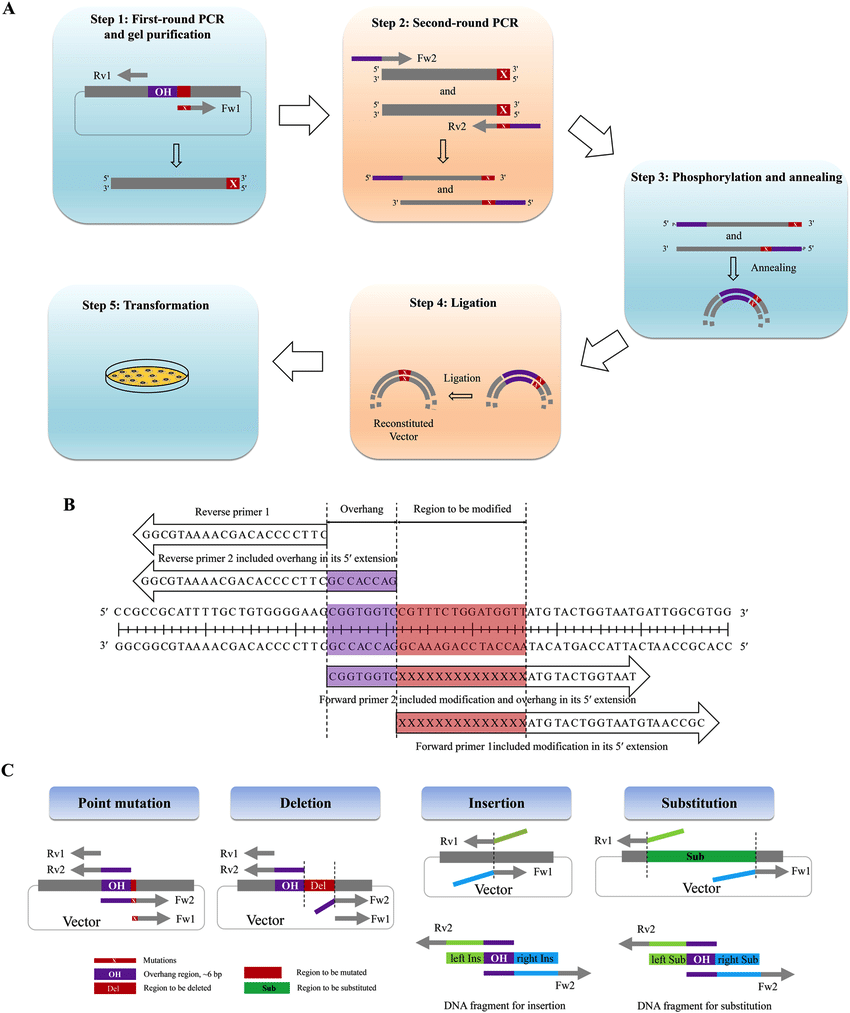
20. **Based on the parameters that affect DNA melting , predict how PCR conditions should be modified to allow the use of a mutagenic primer oligonucleotide**
- AT rich melts earlier / at lower temperatures
- GC rich melts later / at higher temperatures
- melting temp of DNA is proportional to sodium ion concentration
- melting temp is proportional to GC content
- as the length of DNA increases , then melting temperature increases
- if you have a very long DNA molecule , the chances of reannealing are higher
- melting temp is inversely proportional of percent of mismatches
- if you have 3 DNA molecules , 2 are exactly complementary , and 1 is 98% complementary
- the DNA with perfect complementary has a higher melting temperature than the imperfect one
- AA/ TT base pair stacking
- free energy ( energy required to melt ) is pretty stable
- one of the primers your going to use doesn't exactly match the template
- make a primer that overlaps the region you want to change
- 3' end of the primer can still bind to the template
- but there is a mismatch on the 5' end by the primer
- whether the mismatch tail is actually annealed or not , it doesn't matter
- on the next cycle of PCR , the primer will copy the whole thing on the next sequence
<img src="https://39363.org/IMAGE_BUCKET/1663288000183-172825154.jpeg" style="zoom:80%;" />
21. **You would like to clone the cDNA for the enzyme that makes Rudolph the Red Nose Reindeer turn red.**
- **What cells would you use?**
- nose cells
- **What would you isolate?**
- nosagloase mRNA
- **How would you do the cloning?**
- everse transcribe it to cDNA from mRNA of the illuminating protein
- **How would you identify your target clone?**
- look at it , it should glow red
- everything else has all of the other cDNAs from the rest of the library
22. **You would like to clone the human vitamin D receptor which is expressed in all cells of the body**
- **What cells would you use?**
- random gut cells or any random cell
- **What would you isolate?**
- vitamin D receptor mRNA
- **How would you do the cloning?**
- create cDNA from mRNA of the vitamin D receptor mRNA
- **How would you identify your target clone?**
- affinity column chromatography using vitamin D in the beads. Or if you could fluorescently tag vitamin D you could do it in a plate colony
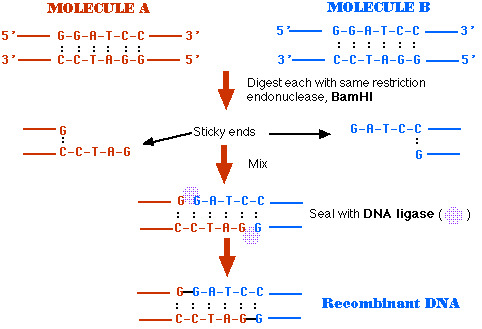
23. **Restriciton enzymes are extensively used in molecular biology. Below are the restriction sites of two of these enzymes , BamH1 and Bcl1**
- a.) = **BamH1 , cleaves after the first $G$. What is the sequence of the sticky end ( overhang ) produced ?**
- CTAG and GATC
<img src="https://39363.org/IMAGE_BUCKET/1663038059147-308927078.png" style="zoom:67%;" />
- b.) = **Bcl1 cleaves after the first $T$. What is the sequence of its overhang?**
- GATC and GATC
<img src="https://39363.org/IMAGE_BUCKET/1663038072703-110025913.png" style="zoom:67%;" />
1. c.) = **You are given the following DNA :**
<img src="https://39363.org/IMAGE_BUCKET/1663038082330-969203359.png" style="zoom:67%;" />
- **If a sample of this DNA was cut BamH1 and another sample was cut with Bcl1 ,**
- **you could ligate the fragments with BamH1 ends to the fragments with Bcl1 ends.**
- **Explain why**
- BamH1 :
- Recognition Sequence :
- 5' GGATCC
- 3' CCTAGG
- Cut :
- 5' ---G <span style="color: green">GATCC--- 3'</span>
- 3' ---CCTAG G--- 5'
- Bcl1 :
- Recognition Sequence :
- 5' TGATCA
- 3' ACTAGT
- Cut :
- 5' ---T GATCA--- 3'
- <span style="color: gold">3' ---ACTAG</span> T--- 5'
- <span style="color: green">5' GATCC--- 3'</span>
- <span style="color: gold">3' ---ACTAG 5'</span>
- **Which enzyme ( BamH1 or Bcl1 ) could be used to cut the ligated fragments from ( c ) ?**
- Neither
- **Explain why**
- just because you've got complementarity of the sticky ends does not necessarily mean that you've got restoration of the entire recognition site.
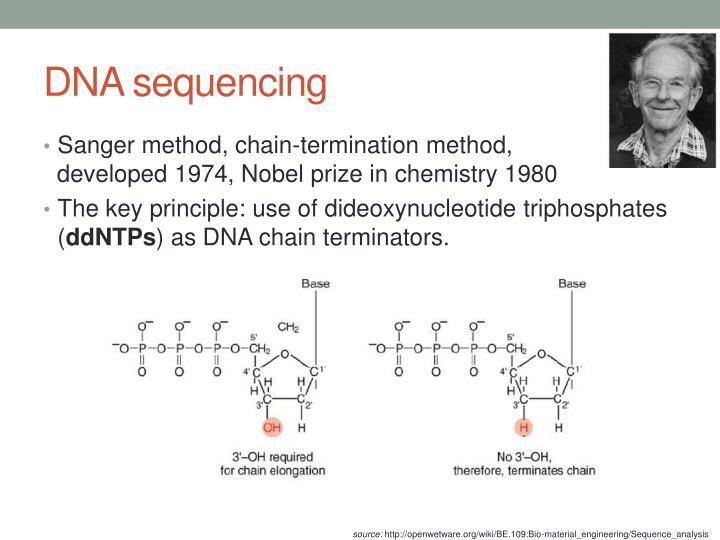
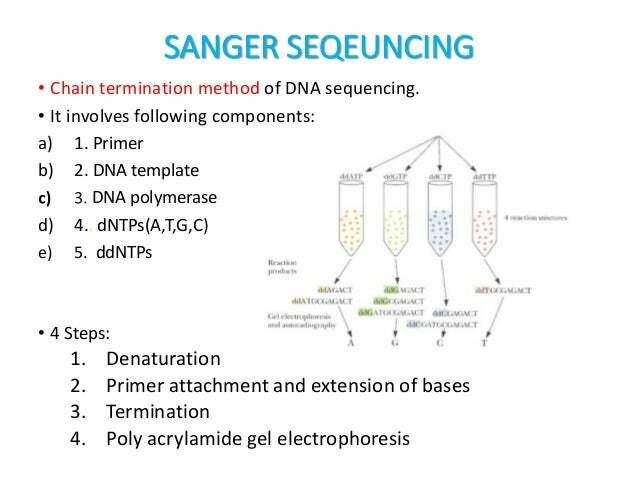
24. **Distinguish between chain termination ( ddNTP ) DNA sequencing and other currently used methods of DNA sequencing**
<img src="https://39363.org/IMAGE_BUCKET/1663293983963-418448639.png" style="zoom:50%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663294013468-85147312.png" style="zoom:50%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663293838283-4854898.jpeg" style="zoom:75%;" />
<img src="https://39363.org/IMAGE_BUCKET/1663294043819-336355797.png" style="zoom:50%;" />
25. **Relate the following assays to DNA annealing :**
- **PCR**
- the second step of PCR is to anneal the primer to the 3' end
- **DNA sequencing**
- first step in Sanger's Sequencing is annealing of a primer to an unknown template strand
- **Southern Blotting**
- The probe is hybridized / annealed onto the strand
<img src="https://39363.org/IMAGE_BUCKET/1663295273874-48339678.png" style="zoom:67%;" />
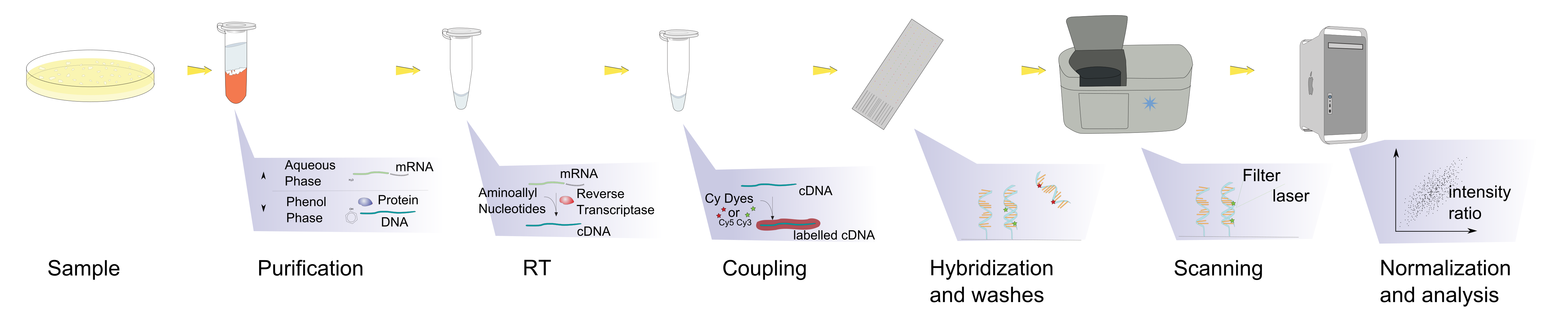
- **Microarray Analysis**
- again in the hybridization step , annealing occurs
26. **How would you calculate the minimum length ( primer length = $L$ ) of a PCR primer oligonucleotide that should have only one specific binding site in a human genome ( $3.3 *10^9$ bp haploid human genome ) ?**
- $3.3 * 10^9 * \left(\ \text{primer length} \ \right) = 1$
- $\frac{3.3 * 10^9}{\left(\ \text{primer length} \ \right)} = 1$
- <span style="border: 2px solid green;">$\frac{3.3 * 10^9}{4^{\left(\ \text{primer length} \ \right)}} = 1$</span>
- $\frac{3.3 * 10^9}{2^{\left(\ \text{primer length} \ \right)}} = 1$
- $4^{\left(\ \text{primer length} \ \right)} = 1$
$$
\frac{3.3 * 10^9}{4^{\left(\ L \ \right)}} = 1
$$
$$
L \approx 15.8099 \approx 16
$$
$$
\frac{3.3 * 10^9}{4^{\left(\ 16 \ \right)}} = 1
$$
- if you have an oligonucleotide of five bases, there will be 3 million sites in theory where it could bind perfectly
27. **(How might RFLP’s be used to monitor the effect of pollutants on the biodiversity of a natural ecosystem?)**
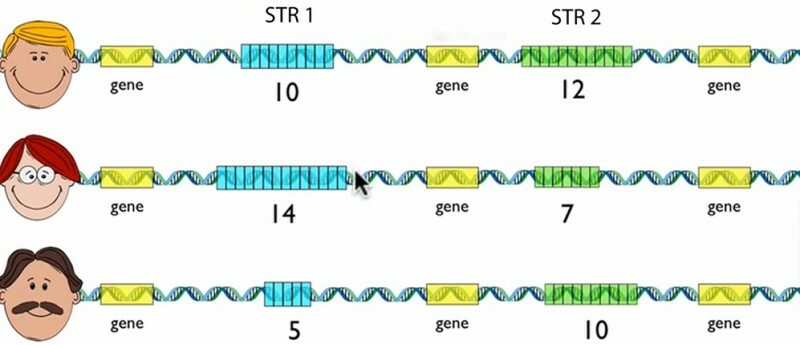
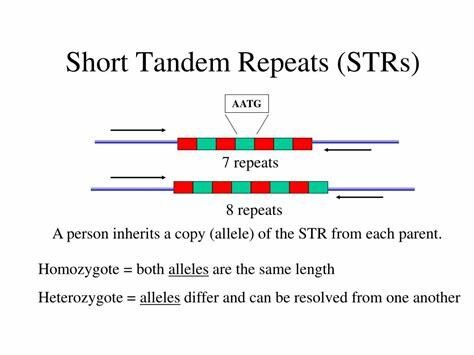
- STR = short tandem repeats
- The number of repeats distinguishes one person from another
- As the population shrinks from pollution , you would expect to see fewer and fewer bands.
- The population is converging to a lineage of only those who can survive pollutants
- Each human inherits one chromosome from each parent ,
- the STR lengths on the two chromosomes are often different ,
- generating two signals from one individual.
- If multiple STR loci are analyzed , a profile can be generated that is essentially unique to a particular individual.
- PCR amplification allows investigators to obtain DNA fingerprints from less than 1 ng of partially degraded DNA, an amount that can be obtained from a single hair follicle, a drop of blood, a small semen sample on a bed sheet, or samples that might be months or even many years old.
28. <img src="https://39363.org/IMAGE_BUCKET/1663038384238-970253970.png" style="zoom:100%;" />
- from Dad , we get only one 2.3 kb restriction fragment
- therefore , Dad does NOT have a polymorphism from this restriction enzyme and probe
- both of his parents gave him a 2.3 kb band
- Mom gives two bands
- which of these bands is linked to a mutant gene , and which is linked to the wild type ?
- If we look at Jane , Jane is affected with the disease
- disease = recessive = so she has 2 mutant copies
- She got the 2.3 band from Mom AND from Dad
- Mom gave her the 2.3 band
- so the 2.3 kb band must be linked in mom to the mutant gene
- Dad gave her the 1.8 and the 2.3
- Did mom give the mutant gene to the fetus ?
- Yes , the fetus is at least a carrier
- We have to look for a probe and enzyme set where Dad DID have a polymorphisms
- Dad gave the 4.8 band to Jane
- he did NOT give the 6.2 band to Jane
- so this band ( 4.8 ) must be on the same chromosome as the mutant gene
- which chromosome did dad give to the fetus ?
- 4.8 kb band
- Mom gave the 2.3 band , and Dad gave the 4.8 band
- so the fetus will be positive for the disease
- **Which of Mom's chromosomes contains the mutant gene ?**
- 2.3 band met H Msp I
- **Which of Dad's chromosomes contains the mutant gene ?**
- 4.8 band met D Taq I
- **What phenotype is predicted for Mary ?**
- Heterozygous for 2.3 met H Msp I
- Wild Type for 4.8 met D Taq I
- **What phenotype is predicted for the Fetus ?**
- Mom gave the 2.3 band , and Dad gave the 4.8 band
- so the fetus will be positive for the disease
29. <img src="https://39363.org/IMAGE_BUCKET/1663038539515-920148588.png" style="zoom:50%;" />
- Dad = Carrier
- Mom = Carrier
- Which band , the 2.3 or the 1.8 did Mom give to David and Sally?
- She gave the 1.8 to david and sally
- because dad does NOT have a 1.8 kb band
- Which band did Mom give to Jane ? and Which band did Mom give to John?
- She have the 2.3 band to Jane and John
- She gave the 1.8 band to John
- Which chromosome did Dad give to David and Sally?
- he gave the 4.2
- must be a mutant chromosome
- Which mutant chromosome did Dad give to Jane and John ?
- Dad must have given the wild type to Jane , but he must of given the mutant to John ?
- Dad gave wild type to Jane
- Jane = normal
- Dad gave mutant to John
- John = carrier
- Whereever the "two marks are" :
- one mark = mutant
- one mark = wild type
-
- **Which of Mom's chromosomes contains the mutant gene ?**
- 4.8 met D Taq I
- **Which of Dad's chromosomes contains the mutant gene ?**
- 1.8 met H MsP I
- 4.0 met H Taq I
- **What phenotype is predicted for Jane ?**
- negative for :
- 1.8 met H Msp I
- 4.2 J 3.11 MsP I
- 4.0 met H Taq I
- **For the John ?**
- negative for :
- 1.8 met H Msp I
- 4.0 met H Taq I
- 6.8 met D Ban I
- 4.8 met D Taq I
30. **What can you say about the phenotype of the fetus ?**
<img src="https://39363.org/IMAGE_BUCKET/1663038605591-652361874.png" style="zoom:67%;" />
- negative for 8 J3.11 Msp I
- negative for 6.3 J3.11 Taq I
-
- Snap-Back DNA = rapidly renaturing DNA fragments due to intramolecular base-pairing
- Satellite DNA =
- PCR
- Restriciton Enzymes
- Nucleotide Metabolism
- DNA sequencing
 Sign in with Wallet
Sign in with Wallet

