[](https://hackmd.io/SAKzGZa0TX-oDKQ6nSd15A)
# Evolution of NLRs: Testing convergence versus history
## Data
## Methods
### Download data from NCBI
We downloaded proteome data from NCBI for 127 different plant species (November 2020). The whole list of proteomes that have been downloaded is here:
```{text}
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/735/GCF_000001735.3_TAIR10/GCF_000001735.3_TAIR10_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/735/GCF_000001735.4_TAIR10.1/GCF_000001735.4_TAIR10.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/002/425/GCF_000002425.3_V1.1/GCF_000002425.3_V1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3218/100/GCF_000002425.4_Phypa_V3/GCF_000002425.4_Phypa_V3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/002/595/GCF_000002595.1_v3.0/GCF_000002595.1_v3.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/002/775/GCF_000002775.3_Poptr2_0/GCF_000002775.3_Poptr2_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3694/101/GCF_000002775.4_Pop_tri_v3/GCF_000002775.4_Pop_tri_v3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/003/195/GCF_000003195.2_Sorbi1/GCF_000003195.2_Sorbi1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4558/101/GCF_000003195.3_Sorghum_bicolor_NCBIv3/GCF_000003195.3_Sorghum_bicolor_NCBIv3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/003/745/GCF_000003745.2_12X/GCF_000003745.2_12X_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/29760/102/GCF_000003745.3_12X/GCF_000003745.3_12X_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/004/075/GCF_000004075.1_CucSat_1.0/GCF_000004075.1_CucSat_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3659/101/GCF_000004075.2_ASM407v2/GCF_000004075.2_ASM407v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3659/102/GCF_000004075.3_Cucumber_9930_V3/GCF_000004075.3_Cucumber_9930_V3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/004/255/GCF_000004255.1_v.1.0/GCF_000004255.1_v.1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/81972/101/GCF_000004255.2_v.1.0/GCF_000004255.2_v.1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/004/515/GCF_000004515.3_V1.1/GCF_000004515.3_V1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/004/515/GCF_000004515.4_Glycine_max_v2.0/GCF_000004515.4_Glycine_max_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3847/103/GCF_000004515.5_Glycine_max_v2.1/GCF_000004515.5_Glycine_max_v2.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/005/GCF_000005005.1_B73_RefGen_v3/GCF_000005005.1_B73_RefGen_v3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4577/102/GCF_000005005.2_B73_RefGen_v4/GCF_000005005.2_B73_RefGen_v4_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/425/GCF_000005425.2_Build_4.0/GCF_000005425.2_Build_4.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/505/GCF_000005505.1_v1.0/GCF_000005505.1_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/005/505/GCF_000005505.2_Brachypodium_distachyon_v2.0/GCF_000005505.2_Brachypodium_distachyon_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/15368/103/GCF_000005505.3_Brachypodium_distachyon_v3.0/GCF_000005505.3_Brachypodium_distachyon_v3.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/090/985/GCF_000090985.2_ASM9098v2/GCF_000090985.2_ASM9098v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/091/205/GCF_000091205.1_ASM9120v1/GCF_000091205.1_ASM9120v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/092/065/GCF_000092065.1_ASM9206v1/GCF_000092065.1_ASM9206v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/143/415/GCF_000143415.3_v1.0/GCF_000143415.3_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/88036/100/GCF_000143415.4_v1.0/GCF_000143415.4_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/143/455/GCF_000143455.1_v1.0/GCF_000143455.1_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/147/415/GCF_000147415.1_v_1.0/GCF_000147415.1_v_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/148/765/GCF_000148765.1_MalDomGD1.0/GCF_000148765.1_MalDomGD1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3649/100/GCF_000150535.2_Papaya1.0/GCF_000150535.2_Papaya1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/151/265/GCF_000151265.1_Micromonas_pusilla_CCMP1545_v2.0/GCF_000151265.1_Micromonas_pusilla_CCMP1545_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/151/265/GCF_000151265.2_Micromonas_pusilla_CCMP1545_v2.0/GCF_000151265.2_Micromonas_pusilla_CCMP1545_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3988/101/GCF_000151685.1_JCVI_RCG_1.1/GCF_000151685.1_JCVI_RCG_1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/57918/101/GCF_000184155.1_FraVesHawaii_1.0/GCF_000184155.1_FraVesHawaii_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/188/115/GCF_000188115.2_SL2.40/GCF_000188115.2_SL2.40_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/188/115/GCF_000188115.3_SL2.50/GCF_000188115.3_SL2.50_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4081/103/GCF_000188115.4_SL3.0/GCF_000188115.4_SL3.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3641/100/GCF_000208745.1_Criollo_cocoa_genome_V2/GCF_000208745.1_Criollo_cocoa_genome_V2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/214/015/GCF_000214015.2_version_050606/GCF_000214015.2_version_050606_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/214/015/GCF_000214015.3_version_140606/GCF_000214015.3_version_140606_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/219/495/GCF_000219495.1_MedtrA17_3.5/GCF_000219495.1_MedtrA17_3.5_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/219/495/GCF_000219495.2_MedtrA17_4.0/GCF_000219495.2_MedtrA17_4.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3880/101/GCF_000219495.3_MedtrA17_4.0/GCF_000219495.3_MedtrA17_4.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/224/045/GCF_000224045.1_CSB10A_v1/GCF_000224045.1_CSB10A_v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4113/101/GCF_000226075.1_SolTub_3.0/GCF_000226075.1_SolTub_3.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4533/101/GCF_000231095.1_Oryza_brachyantha.v1.4b/GCF_000231095.1_Oryza_brachyantha.v1.4b_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/258/705/GCF_000258705.1_Coccomyxa_subellipsoidae_v2.0/GCF_000258705.1_Coccomyxa_subellipsoidae_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/263/155/GCF_000263155.1_Setaria_V1/GCF_000263155.1_Setaria_V1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4555/103/GCF_000263155.2_Setaria_italica_v2.0/GCF_000263155.2_Setaria_italica_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3711/101/GCF_000309985.1_Brapa_1.0/GCF_000309985.1_Brapa_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3656/101/GCF_000313045.1_ASM31304v1/GCF_000313045.1_ASM31304v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/313/855/GCF_000313855.1_ASM31385v1/GCF_000313855.1_ASM31385v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4641/101/GCF_000313855.2_ASM31385v2/GCF_000313855.2_ASM31385v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/225117/101/GCF_000315295.1_Pbr_v1.0/GCF_000315295.1_Pbr_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/2711/102/GCF_000317415.1_Csi_valencia_1.0/GCF_000317415.1_Csi_valencia_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/29730/100/GCF_000327365.1_Graimondii2_0/GCF_000327365.1_Graimondii2_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3827/102/GCF_000331145.1_ASM33114v1/GCF_000331145.1_ASM33114v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3821/101/GCF_000340665.1_C.cajan_V1.0/GCF_000340665.1_C.cajan_V1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/341/285/GCF_000341285.1_ASM34128v1/GCF_000341285.1_ASM34128v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/346/465/GCF_000346465.1_Prupe1_0/GCF_000346465.1_Prupe1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3760/100/GCF_000346465.2_Prunus_persica_NCBIv2/GCF_000346465.2_Prunus_persica_NCBIv2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/102107/101/GCF_000346735.1_P.mume_V1.0/GCF_000346735.1_P.mume_V1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/350/225/GCF_000350225.1_ASM35022v2/GCF_000350225.1_ASM35022v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4432/101/GCF_000365185.1_Chinese_Lotus_1.1/GCF_000365185.1_Chinese_Lotus_1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/81985/100/GCF_000375325.1_Caprub1_0/GCF_000375325.1_Caprub1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4098/101/GCF_000390325.1_Ntom_v01/GCF_000390325.1_Ntom_v01_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/390/325/GCF_000390325.2_Ntom_v01/GCF_000390325.2_Ntom_v01_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4096/100/GCF_000393655.1_Nsyl/GCF_000393655.1_Nsyl_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/403/535/GCF_000403535.1_Theobroma_cacao_20110822/GCF_000403535.1_Theobroma_cacao_20110822_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/42345/102/GCF_000413155.1_DPV01/GCF_000413155.1_DPV01_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/981085/100/GCF_000414095.1_ASM41409v2/GCF_000414095.1_ASM41409v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/51953/102/GCF_000442705.1_EG5/GCF_000442705.1_EG5_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/28532/101/GCF_000463585.1_ASM46358v1/GCF_000463585.1_ASM46358v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/471/905/GCF_000471905.1_AMTR1.0/GCF_000471905.1_AMTR1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/13333/101/GCF_000471905.2_AMTR1.0/GCF_000471905.2_AMTR1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/72664/100/GCF_000478725.1_Eutsalg1_0/GCF_000478725.1_Eutsalg1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/85681/100/GCF_000493195.1_Citrus_clementina_v1.0/GCF_000493195.1_Citrus_clementina_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/75702/100/GCF_000495115.1_PopEup_1.0/GCF_000495115.1_PopEup_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/499/845/GCF_000499845.1_PhaVulg1_0/GCF_000499845.1_PhaVulg1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4155/100/GCF_000504015.1_Mimgu1_0/GCF_000504015.1_Mimgu1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/511/025/GCF_000511025.1_RefBeet-1.2.1/GCF_000511025.1_RefBeet-1.2.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3555/101/GCF_000511025.2_RefBeet-1.2.2/GCF_000511025.2_RefBeet-1.2.2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4182/101/GCF_000512975.1_S_indicum_v1.0/GCF_000512975.1_S_indicum_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/611/645/GCF_000611645.1_mono_v1/GCF_000611645.1_mono_v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/29729/100/GCF_000612285.1_Gossypium_arboreum_v1.0/GCF_000612285.1_Gossypium_arboreum_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/71139/101/GCF_000612305.1_Egrandis1_0/GCF_000612305.1_Egrandis1_0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/90675/101/GCF_000633955.1_Cs/GCF_000633955.1_Cs_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/686/985/GCF_000686985.1_Brassica_napus_assembly_v1.0/GCF_000686985.1_Brassica_napus_assembly_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3708/101/GCF_000686985.2_Bra_napus_v2.0/GCF_000686985.2_Bra_napus_v2.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3712/100/GCF_000695525.1_BOL/GCF_000695525.1_BOL_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/180498/101/GCF_000696525.1_JatCur_1.0/GCF_000696525.1_JatCur_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4072/100/GCF_000710875.1_Pepper_Zunla_1_Ref_v1.0/GCF_000710875.1_Pepper_Zunla_1_Ref_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4097/100/GCF_000715135.1_Ntab-TN90/GCF_000715135.1_Ntab-TN90_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/733/215/GCF_000733215.1_ASM73321v1/GCF_000733215.1_ASM73321v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/157791/101/GCF_000741045.1_Vradiata_ver6/GCF_000741045.1_Vradiata_ver6_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3726/100/GCF_000801105.1_Rs1.0/GCF_000801105.1_Rs1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/816/755/GCF_000816755.1_Araip1.0/GCF_000816755.1_Araip1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/130454/101/GCF_000816755.2_Araip1.1/GCF_000816755.2_Araip1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/817/695/GCF_000817695.1_Aradu1.0/GCF_000817695.1_Aradu1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/130453/101/GCF_000817695.2_Aradu1.1/GCF_000817695.2_Aradu1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/326968/101/GCF_000826755.1_ZizJuj_1.1/GCF_000826755.1_ZizJuj_1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3635/100/GCF_000987745.1_ASM98774v1/GCF_000987745.1_ASM98774v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3914/100/GCF_001190045.1_Vigan1.1/GCF_001190045.1_Vigan1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/78828/100/GCF_001263595.1_ASM126359v1/GCF_001263595.1_ASM126359v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/28526/101/GCF_001406875.1_SPENNV200/GCF_001406875.1_SPENNV200_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/51240/100/GCF_001411555.1_wgs.5d/GCF_001411555.1_wgs.5d_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/39947/102/GCF_001433935.1_IRGSP-1.0/GCF_001433935.1_IRGSP-1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/309979/100/GCF_001531365.1_CcrdV1/GCF_001531365.1_CcrdV1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4615/100/GCF_001540865.1_ASM154086v1/GCF_001540865.1_ASM154086v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/142615/100/GCF_001605985.1_ASM160598v1/GCF_001605985.1_ASM160598v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/906689/101/GCF_001605985.2_ASM160598v2/GCF_001605985.2_ASM160598v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/79200/100/GCF_001625215.1_ASM162521v1/GCF_001625215.1_ASM162521v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/97700/100/GCF_001633185.1_ValleyOak3.0/GCF_001633185.1_ValleyOak3.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3981/100/GCF_001654055.1_ASM165405v1/GCF_001654055.1_ASM165405v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3983/100/GCF_001659605.1_Manihot_esculenta_v6/GCF_001659605.1_Manihot_esculenta_v6_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/63459/100/GCF_001683475.1_ASM168347v1/GCF_001683475.1_ASM168347v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3871/100/GCF_001865875.1_LupAngTanjil_v1.0/GCF_001865875.1_LupAngTanjil_v1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4686/100/GCF_001876935.1_Aspof.V1/GCF_001876935.1_Aspof.V1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/49451/100/GCF_001879085.1_NIATTr2/GCF_001879085.1_NIATTr2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/35883/100/GCF_001879475.1_Asagao_1.1/GCF_001879475.1_Asagao_1.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/37682/100/GCF_001957025.1_Aet_MR_1.0/GCF_001957025.1_Aet_MR_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3673/100/GCF_001995035.1_ASM199503v1/GCF_001995035.1_ASM199503v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3562/100/GCF_002007265.1_ASM200726v1/GCF_002007265.1_ASM200726v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3750/102/GCF_002114115.1_ASM211411v1/GCF_002114115.1_ASM211411v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4232/100/GCF_002127325.1_HanXRQr1.0/GCF_002127325.1_HanXRQr1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/108875/100/GCF_002168275.1_ASM216827v2/GCF_002168275.1_ASM216827v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/42229/100/GCF_002207925.1_PAV_r1.0/GCF_002207925.1_PAV_r1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/206008/100/GCF_002211085.1_PHallii_v3.1/GCF_002211085.1_PHallii_v3.1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/002/220/235/GCF_002220235.1_ASM222023v1/GCF_002220235.1_ASM222023v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/66656/100/GCF_002303985.1_Duzib1.0/GCF_002303985.1_Duzib1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3661/100/GCF_002738345.1_Cmax_1.0/GCF_002738345.1_Cmax_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3662/100/GCF_002738365.1_Cmos_1.0/GCF_002738365.1_Cmos_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/158386/100/GCF_002742605.1_O_europaea_v1/GCF_002742605.1_O_europaea_v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3664/100/GCF_002806865.1_ASM280686v2/GCF_002806865.1_ASM280686v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4236/100/GCF_002870075.1_Lsat_Salinas_v7/GCF_002870075.1_Lsat_Salinas_v7_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/58331/100/GCF_002906115.1_CorkOak1.0/GCF_002906115.1_CorkOak1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/74649/100/GCF_002994745.1_RchiOBHm-V2/GCF_002994745.1_RchiOBHm-V2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/003/086/295/GCF_003086295.1_arahy.Tifrunner.gnm1.KYV3/GCF_003086295.1_arahy.Tifrunner.gnm1.KYV3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3818/101/GCF_003086295.2_arahy.Tifrunner.gnm1.KYV3/GCF_003086295.2_arahy.Tifrunner.gnm1.KYV3_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3469/100/GCF_003573695.1_ASM357369v1/GCF_003573695.1_ASM357369v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/35885/100/GCF_003576645.1_ASM357664v1/GCF_003576645.1_ASM357664v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/49369/100/GCF_003713205.1_Ceug_1.0/GCF_003713205.1_Ceug_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/13443/100/GCF_003713225.1_Cara_1.0/GCF_003713225.1_Cara_1.0_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3816/100/GCF_003935025.1_Abrus_2018/GCF_003935025.1_Abrus_2018_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3917/100/GCF_004118075.1_ASM411807v1/GCF_004118075.1_ASM411807v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/4442/100/GCF_004153795.1_AHAU_CSS_1/GCF_004153795.1_AHAU_CSS_1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3848/100/GCF_004193775.1_ASM419377v2/GCF_004193775.1_ASM419377v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/207710/100/GCF_004799145.1_ASM479914v1/GCF_004799145.1_ASM479914v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/22663/100/GCF_007655135.1_ASM765513v2/GCF_007655135.1_ASM765513v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/55513/100/GCF_008641045.1_PisVer_v2/GCF_008641045.1_PisVer_v2_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/210225/100/GCF_008831285.1_ASM883128v1/GCF_008831285.1_ASM883128v1_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/3483/100/GCF_900626175.1_cs10/GCF_900626175.1_cs10_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/178133/100/GCF_900635035.1_Rarg10x-PRISCAF/GCF_900635035.1_Rarg10x-PRISCAF_protein.faa.gz
ftp://ftp.ncbi.nlm.nih.gov/genomes/all/annotation_releases/219896/100/GCF_900635055.1_Sole10x-PRISCAF/GCF_900635055.1_Sole10x-PRISCAF_protein.faa.gz
```
From this list, some of the files refer to **the same** proteome. Thus, for those files that refer to the same proteome ,we kept only the file with the greatest index number, i.e, the most recent version of the proteome.
### Run the NLR detection pipeline
We used perl custom scripts to run automatically the analysis presented by Sarris et al. (2016).
Aiming to annotate domains and extract NLR plant immune receptors and their architectures, we followed the steps described in a set of scripts which is available from GitHub (https://github.com/krasileva/plant_r-genes). The output is a summary of the number of NLRs and NLR integrated domains identified in each species, along with a summary of integrated domains with species list for each domain. A large list of integrated domains counted once for each family and contingency tables per integrated domain, for each species as well as for all species and Fisher’s Exact left test.
Reciprocal BLAST analysis for orthologue identification and phylogenetic tree reconstruction from genetic data.
The main script (`cmds_externalName_rename.sh`) employed to perform the NLR detection is the following:
```{bash}
## THIS IS the main execution script
## to extract the NLRs from PROTEOMES
## IT also renames the files
## PLEASE CHANGE THE PATHS IF YOU NEED TO RUN IT IN YOUR COMPUTER
PATH=$PATH:~/synology/tools/nlr_detection/bin
export PATH
HMMERDB=~/synology/PFAM/
export HMMERDB
curDir=`pwd`
file=$1
renameFile=`basename $file .faa.gz`
echo "RENAME $file to $renameFile" > $file.RENAME
ln -s $file $renameFile.fasta.gz
file=$renameFile
echo $file
mkdir $file
cd $file
## make the database
make_db.pl $file > db_description.txt
## rename the data
gunzip -c ../$file.fasta.gz > $file.fasta
ln -s $file.fasta $file.protein.fa
run_pfam_scan.sh ./
ls ./pfam/$file.protein*.out
inputFile=`ls ./pfam/$file.protein.*.out`
echo $inputFile
K-parse_Pfam_domains_v3.1.pl -p $inputFile -e 0.001 -o $inputFile.parsed.verbose -v T
K-parse_Pfam_domains_NLR-fusions-v2.2.pl -i ./pfam -e 0.001 -o ./pfam -d db_description.txt
cd $curDir
```
### Detection of orthologs, alignment, supermatrix and ML phylogenetic tree
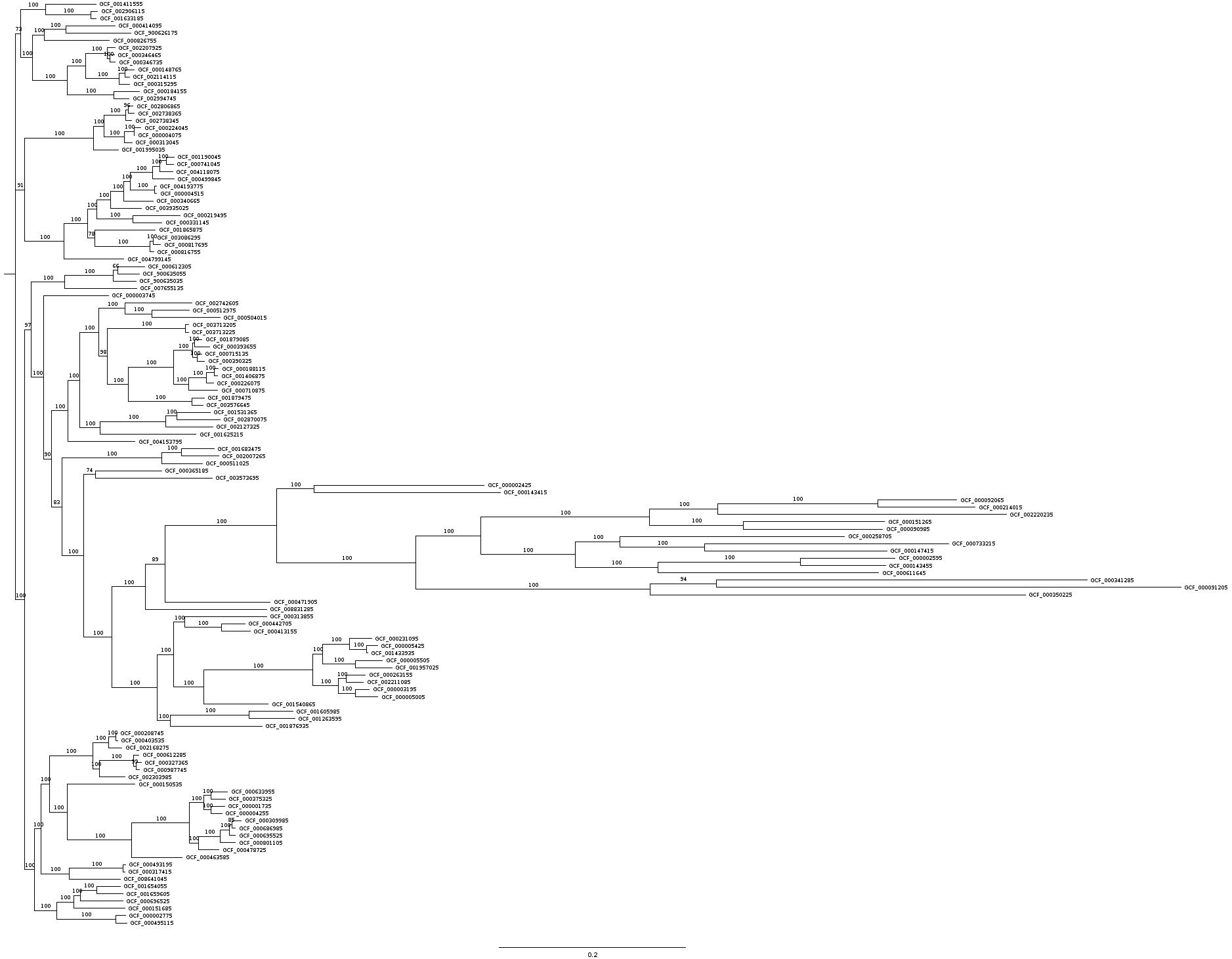
For each of the 126 plant proteomes we performed a reciprocal BLAST analysis against the A.thaliana proteome to identify orthologous protein sequences. The parameters of the blastp program were set to `-outfmt 6`, `-evalue 1e-10` and `-num_alignments 1` to obtain only the best matched protein. Given that the best hit reciprocally fulfills the e-value BLAST criterion (1x10-10) then the orthology is assigned. Eventually, a set of 481 proteins was obtained, which fulfill the aforementioned criteria for all species. These 481 proteins were aligned using mafft (`--maxiterate 1000 --localpair`) and then merged with a custom perl script to generate a supermatrix alignment. Finally, raxml-ng (`--model WAG --tree pars{20} -bs-trees 100`) was employed to reconstruct the phylogenetic tree for the protein data (Figure XXXX)
The following scripts were used to perform the reciprocal BLAST analysis, build the supermatrix and reconstruct the phylogeny (see folder /home/zoe/synology/zoe/analysis/separate_blast in Maya).
#### Script 1 -- Construct the blast databases for all plant proteomes
```{bash}
zoe@Maya:~/synology/zoe/analysis/separate_blast$ more make_blast_all.sh
ls *.protein.fa > fa.list
for f in `cat fa.list`;do
makeblastdb -in $f -parse_seqids -title $f -dbtype prot
done
```
#### Script 2 -- Run the blast analysis
```{bash}
zoe@Maya:~/synology/zoe/analysis/separate_blast$ more run_blast.sh
threads=16
for f in `cat fa.list`;do
blastp -num_threads $threads -query GCF_000001735.4_TAIR10.1_protein.protein.fa -db $f -num_alignments 1 -outfmt 6 -evalue 1e-10 -ou
t TAIR_to_${f}.blastres
blastp -num_threads $threads -query $f -db GCF_000001735.4_TAIR10.1_protein.protein.fa -num_alignments 1 -outfmt 6 -evalue 1e-10 -ou
t ${f}_to_TAIR.blastres
echo $f
done
```
#### Script 3 -- Analyze blast results to keep only the orthologues
```{bash}
ls GCF*.blastres | sed 's/_to_TAIR.*//' > tmp.list
for f in `cat tmp.list`;do
g=`basename $f .blastres`;
./bin/parse_reciprocal_blast.pl -1 TAIR_to_${f}.blastres -2 ${f}_to_TAIR.blastres > ${f}.goodres;
done
ls *.goodres | grep -v blastres | grep -v _.goodres > goodres.list
wc -l goodres.list
./bin/count_blast_hits_common.pl -ref TAIR.genes -blast goodres.list -n 127
```
The last script uses a perl script to parse BLAST results.
#### Perl script to parse BLAST results
```{perl}
#!/usr/bin/perl -w
use strict;
### LOCATION: /home/zoe/synology/zoe/analysis/separate_blast/bin
my $usage = "It parses the results of reciprocal blast to find the pairs that match the criterion of reciprocality\n./parse_reciprocal_blast.pl -1 <FILE 1> -2 <FILE 2>\n\n";
if($#ARGV < 0){ die $usage; }
my $f1 = "";
my $f2 = "";
while( my $args = shift @ARGV){
if( $args =~ /^-1$/i){ $f1 = shift @ARGV; next; }
if( $args =~ /^-2$/i){ $f2 = shift @ARGV; next; }
die "Argument $args is not valid\n\n$usage\n";
}
open(IN1, "$f1") or die "Couldn't open $f1 for input\n";
my $key = "";
my %keys1 = ();
while( defined(my $ln=<IN1>) ){
if($ln=~/([^\s]+)\s+([^\s]+)/){
my $first = $1;
my $second = $2;
my $tmp = $first;
if( $first gt $second){
$tmp = $first;
$first = $second;
$second = $tmp;
}
$key = $first.$second;
$keys1{$key} = 1;
}
}
close IN1;
open(IN2, "$f2") or die "Couldnt open $f2 for input\n";
while( defined(my $ln=<IN2>) ){
if($ln=~/([^\s]+)\s+([^\s]+)/){
my $first = $1;
my $second = $2;
my $tmp = $first;
if( $first gt $second){
$tmp = $first;
$first = $second;
$second = $tmp;
}
$key = $first.$second;
if(defined( $keys1{$key} )){
print $first, "\t", $second, "\t", $key, "\n";
}
}
}
```
The output of the last script provides the pairs of protein names, **one from A. thaliana** and **one from another species** that are marked as `orthologues` (perhaps the term is not very proper since we don't really perform a strict orthologue analysis, but it's proper for our purpose).
```{text}
### the first column: A.thaliana
### the second column: Another species
### the third column: key composed by the concatenation of the previous two columns
NP_178975.1 XP_027060738.1 NP_178975.1XP_027060738.1
NP_196767.1 XP_027060748.1 NP_196767.1XP_027060748.1
NP_567404.1 XP_027060779.1 NP_567404.1XP_027060779.1
NP_172003.1 XP_027060798.1 NP_172003.1XP_027060798.1
NP_197188.1 XP_027060877.1 NP_197188.1XP_027060877.1
NP_188827.1 XP_027060893.1 NP_188827.1XP_027060893.1
NP_179201.1 XP_027060917.1 NP_179201.1XP_027060917.1
NP_001329572.1 XP_027060921.1 NP_001329572.1XP_027060921.1
NP_189605.1 XP_027060926.1 NP_189605.1XP_027060926.1
NP_565486.1 XP_027060943.1 NP_565486.1XP_027060943.1
NP_172118.1 XP_027060975.1 NP_172118.1XP_027060975.1
NP_181804.1 XP_027060982.1 NP_181804.1XP_027060982.1
NP_191374.2 XP_027061003.1 NP_191374.2XP_027061003.1
NP_194543.1 XP_027061010.1 NP_194543.1XP_027061010.1
NP_001326343.1 XP_027061015.1 NP_001326343.1XP_027061015.1
NP_200370.1 XP_027061040.1 NP_200370.1XP_027061040.1
NP_200433.1 XP_027061052.1 NP_200433.1XP_027061052.1
NP_178039.1 XP_027061061.1 NP_178039.1XP_027061061.1
NP_192421.1 XP_027061066.1 NP_192421.1XP_027061066.1
NP_974313.1 XP_027061068.1 NP_974313.1XP_027061068.1
```
The following tree in newick format represents the phylogenetic tree obtained from the ML analysis (with bootstrap support values).
```{text}
(((GCF_001411555:0.054240,(GCF_002906115:0.007329,GCF_001633185:0.005805)100:0.048631)100:0.026638,(((GCF_000414095:0.052818,GCF_900626175:0.069763)100:0.022641,GCF_000826755:0.069598)100:0.012772,(((GCF_002207925:0.008182,(GCF_000346465:0.004870,GCF_000346735:0.006081)100:0.002564)100:0.023513,((GCF_000148765:0.009896,GCF_002114115:0.004841)100:0.006229,GCF_000315295:0.011214)100:0.036236)100:0.019611,(GCF_000184155:0.027840,GCF_002994745:0.016704)100:0.049945)100:0.036891)100:0.012994)73:0.005305,(((((GCF_002806865:0.005263,GCF_002738365:0.006819)96:0.002439,GCF_002738345:0.006464)100:0.023540,((GCF_000224045:0.007245,GCF_000004075:0.000249)100:0.010231,GCF_000313045:0.007949)100:0.022219)100:0.011103,GCF_001995035:0.026738)100:0.073597,(((((((((GCF_001190045:0.008720,GCF_000741045:0.008047)100:0.006447,GCF_004118075:0.013773)100:0.007978,GCF_000499845:0.022859)100:0.024122,(GCF_004193775:0.001515,GCF_000004515:0.002099)100:0.026512)100:0.006758,GCF_000340665:0.031300)100:0.014094,GCF_003935025:0.033252)100:0.014913,(GCF_000219495:0.050595,GCF_000331145:0.031142)100:0.038667)100:0.009990,(GCF_001865875:0.064755,((GCF_003086295:0.000552,GCF_000817695:0.008072)100:0.003079,GCF_000816755:0.003904)100:0.059182)78:0.008001)100:0.024961,GCF_004799145:0.063940)100:0.042158)91:0.009831,(((((GCF_000612305:0.028736,GCF_900635055:0.023012)66:0.005008,GCF_900635035:0.024586)100:0.051588,GCF_007655135:0.076846)100:0.036332,(GCF_000003745:0.069504,(((((GCF_002742605:0.071725,(GCF_000512975:0.039925,GCF_000504015:0.072774)100:0.029043)100:0.027752,((GCF_003713205:0.004042,GCF_003713225:0.003591)100:0.083162,((((GCF_001879085:0.007814,GCF_000393655:0.015985)100:0.001985,(GCF_000715135:0.005016,GCF_000390325:0.008115)100:0.004342)100:0.020582,(((GCF_000188115:0.004514,GCF_001406875:0.003663)100:0.008544,GCF_000226075:0.007735)100:0.018595,GCF_000710875:0.030861)100:0.016368)100:0.048830,(GCF_001879475:0.012773,GCF_003576645:0.011562)100:0.068742)100:0.022014)98:0.009190)100:0.020830,(((GCF_001531365:0.035453,GCF_002870075:0.046003)100:0.011932,GCF_002127325:0.050497)100:0.070312,GCF_001625215:0.102844)100:0.022099)100:0.012401,GCF_004153795:0.071684)100:0.017970,(((GCF_001683475:0.035118,GCF_002007265:0.039934)100:0.020825,GCF_000511025:0.043206)100:0.107398,((GCF_000365185:0.070987,GCF_003573695:0.124936)74:0.012764,(((((GCF_000002425:0.182104,GCF_000143415:0.199422)100:0.040037,(((((GCF_000092065:0.084831,GCF_000214015:0.104496)100:0.171140,GCF_002220235:0.309206)100:0.073233,(GCF_000151265:0.151634,GCF_000090985:0.149373)100:0.100328)100:0.181001,((GCF_000258705:0.240436,(GCF_000733215:0.261780,GCF_000147415:0.195510)100:0.090282)100:0.048170,((GCF_000002595:0.101639,GCF_000143455:0.091434)100:0.152618,GCF_000611645:0.236807)100:0.088607)100:0.101275)100:0.069496,((GCF_000341285:0.397287,GCF_000091205:0.497598)94:0.071105,GCF_000350225:0.402054)100:0.251043)100:0.148957)100:0.119630,GCF_000471905:0.112538)89:0.021558,GCF_008831285:0.130315)100:0.035689,(((GCF_000313855:0.087394,(GCF_000442705:0.025533,GCF_000413155:0.030916)100:0.038848)100:0.012382,((((GCF_000231095:0.023611,(GCF_000005425:0.011810,GCF_001433935:0.001155)100:0.018150)100:0.028677,(GCF_000005505:0.029442,GCF_001957025:0.039382)100:0.034596)100:0.010729,((GCF_000263155:0.020354,GCF_002211085:0.018433)100:0.008811,(GCF_000003195:0.015098,GCF_000005005:0.024137)100:0.018430)100:0.027092)100:0.116917,GCF_001540865:0.069154)100:0.032417)100:0.017573,((GCF_001605985:0.046809,GCF_001263595:0.049037)100:0.084375,GCF_001876935:0.098252)100:0.014375)100:0.048536)100:0.030235)100:0.023369)83:0.011234)90:0.008356)100:0.013359)97:0.006796,((((((GCF_000208745:0.001392,GCF_000403535:0.002139)100:0.007645,GCF_002168275:0.014308)100:0.016824,((GCF_000612285:0.006036,(GCF_000327365:0.005686,GCF_000987745:0.004056)99:0.002848)100:0.036610,GCF_002303985:0.027503)100:0.006917)100:0.046594,(GCF_000150535:0.072403,((((GCF_000633955:0.017920,GCF_000375325:0.015740)100:0.007336,(GCF_000001735:0.014506,GCF_000004255:0.011461)100:0.007790)100:0.015767,((((GCF_000309985:0.009706,GCF_000686985:0.003468)85:0.003314,GCF_000695525:0.006818)100:0.009391,GCF_000801105:0.016083)100:0.023426,GCF_000478725:0.021991)100:0.010083)100:0.061421,GCF_000463585:0.053929)100:0.068931)100:0.019626)100:0.008919,((GCF_000493195:0.002826,GCF_000317415:0.001985)100:0.057311,GCF_008641045:0.054460)100:0.030585)100:0.006776,((((GCF_001654055:0.024840,GCF_001659605:0.027713)100:0.018040,GCF_000696525:0.045806)100:0.006776,GCF_000151685:0.054747)100:0.018426,(GCF_000002775:0.010313,GCF_000495115:0.011879)100:0.063528)100:0.023578)100:0.010650)100:0.009700)73:0.0;
```
## Phylogenetic tree reconstruction from ecological and pathogen data
## Obtaining pathogen data
## Script 1: Get pathogen data
```{r}
#necessary libraries ----
library('devtools')
library('usethis')
library("rglobi")
library("stringr")
library("data.table")
library("doParallel")
library("dplyr")
library("phonTools")
#download data for 127 species ----
setwd("/Users/Zoe/Desktop/ptuxiaki")
species<-read.csv( file="list_final.csv", header = T )
micro<- list()
l<-nrow(species)
for (i in 1:l) {
spe<-species[i,]
print(spe)
micro[[i]]<-get_interactions(taxon = spe , interaction.type = "hasPathogen", otherkeys=list('limit'=2048))
}
tmp<-do.call(rbind, micro)
write.csv(tmp, "/Users/Zoe/Desktop/ptuxiaki/pathogens.csv", row.names = F)
example <- micro[[4]] #find dataframe for specific species in list "micro"
#Species x Pathogen matrix ----
pathogens<-read.csv( file="pathogens.csv", header= T)
uni_pathogens<-unique(tmp[c("target_taxon_name")])
uni_species<-unique(tmp[c("source_taxon_name")])
mtx2<-zeros(nrow(species),nrow(uni_pathogens))
rownames(mtx2)<- species[,1]
colnames(mtx2)<- uni_pathogens[,1]
for ( j in 1:length(micro) ) {
if(nrow(micro[[j]])!=0){
for ( i in 1:nrow(micro[[j]])) {
test_str <- micro[[j]]$target_taxon_name[i]
if(!is.null(test_str)){
mtx2[j,test_str]=1
}
}
}
}
write.csv(mtx2, file="eco_matrix.csv")
```
**TODO:** To classify the 127 species based on their ecological traits, we manually retrieved the information from databases (see the list of databases in the final chapter of this thesis). Thus, creating a dataframe (Supplemental Data/ Eco_traits.csv) containing every species’ ecological profile. Whenever information about a certain trait could not be found, its cell in the dataframe remained empty (and later marked with the appropriate symbol, e.g., a ‘?’ in BEAST analysis).. Since each species is accustomed to a different set of pathogens, we created a similar dataframe with the pathogen list of every species (Supplemental Data/ pathogens.csv ) by using the R package rglobi (Supplemental Data/ get_pathogen_data.R). Subsequently a large matrix was created in which every pathogen was matched to every species (Supplemental Data/ eco_matrix.csv) . A match (i.e., pathogen infects was symbolized with a “1” and a mismatch with a “0”. { Missing text: tree reconstruction from non-binary data }
### Tree of ecological features
#### Description of the data
number of columns and short table with the column names

### How well fit NLR data to genetic data?
### How well fit NLR data to pathogen data?
### How well fit NLR data to ecological data?
 Sign in with Wallet
Sign in with Wallet

