# How to build an application ontology with Robot
**identifier:**
**version:** [v0.1](v0.1)
___
**_Difficulty level:_** :triangular_flag_on_post: :triangular_flag_on_post: :triangular_flag_on_post: :white_circle: :white_circle:
**_Reading time:_** 60 minutes
**_Intended Audience:_**
> :heavy_check_mark: *Terminology Manager*
> :heavy_check_mark: *Ontologist*
> :heavy_check_mark: *Application Developer*
> :heavy_check_mark: *Data Scientist*
**_Recipe Type_**: Hands-on practical
**_Executable code_**: No
---
# How to build application ontology with ROBOT
1. [Main FAIRification Objectives](#Main%20FAIRification%20Objectives)
2. [Graphical Overview of the FAIRification Recipe Objectives](#Graphical%20Overview%20of%20the%20FAIRification%20Recipe%20Objectives)
3. [FAIRification Objectives, Inputs and Outputs](#FAIRification%20Objectives,%20Inputs%20and%20Outputs)
4. [Capability & Maturity Table](#Capability%20&%20Maturity%20Table)
5. [Table of Data Standards](#Table%20of%20Data%20Standards)
6. [Executable Code in Notebook](#Executable%20Code%20in%20Notebook)
7. [How to create workflow figures](#How%20to%20create%20workflow%20figures)
8. [License](#License)
## Main Objectives
> The main purpose of this recipe is building an application ontology from source ontologies using `ROBOT` via a sustainable dynamic pipeline to allow seamless integration of source ontology updates.
An application ontology is a semantic artefact which is developed to answer the needs of a specific application or focus. Thus it may borrow terms from a number of reference ontologies, which can be extremely large but whose broad coverage may not be required by the application ontology. Yet, it is critical to keep the `application ontology` synchronized with the `reference ontologies` that imports are made from. We aim to document how a certain level of automation can be achieved.
[ROBOT](http://robot.obolibrary.org/) is an open source tool for ontology development. It supports ontology editing, ontology annotation, ontology format conversion, and other functions. It runs on the Java Virtual Machine (JVM) and can be used through command line tools on Windows, macOS and Linux.
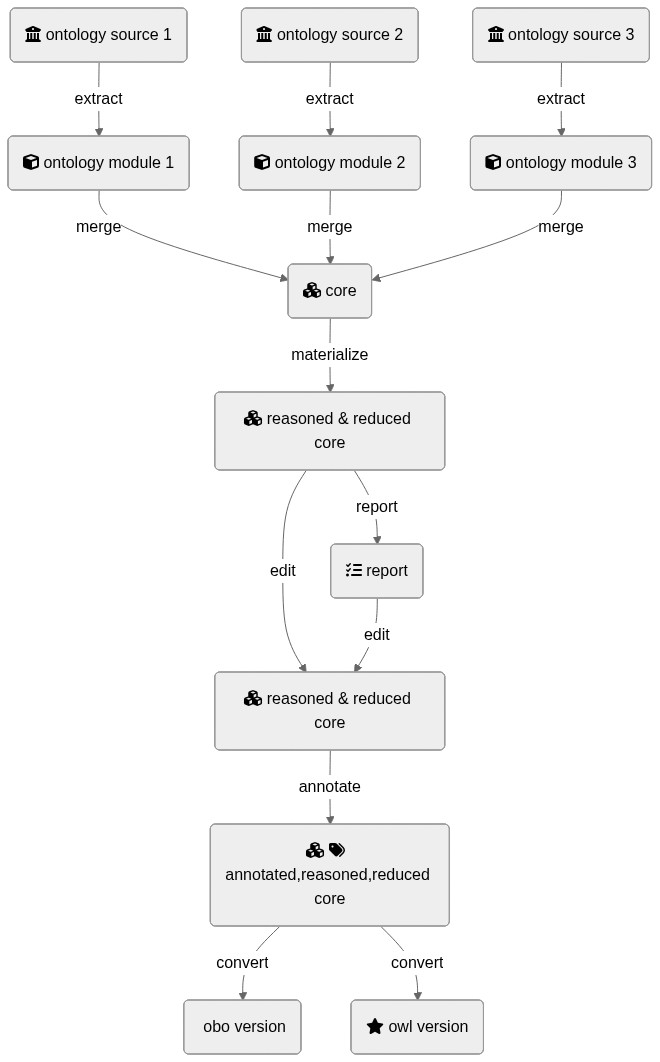
## Graphical Overview of the FAIRification Recipe Objectives
<div class="mermaid">
graph TD
I1(fa:fa-university ontology source 1):::box -->|extract| M1(fa:fa-cube ontology module 1):::box
I2(fa:fa-university ontology source 2):::box -->|extract| M2(fa:fa-cube ontology module 2):::box
I3(fa:fa-university ontology source 3):::box -->|extract| M3(fa:fa-cube ontology module 3):::box
M1 --> |merge| R1(fa:fa-cubes core):::box
M2 --> |merge| R1(fa:fa-cubes core):::box
M3 --> |merge| R1(fa:fa-cubes core):::box
R1(fa:fa-cubes core):::box --> |materialize| R2(fa:fa-cubes reasoned & reduced core):::box
R2(fa:fa-cubes reasoned & reduced core) -->|report| R3(fa:fa-tasks report):::box
R2(fa:fa-cubes reasoned & reduced core) --> |edit| R4(fa:fa-cubes reasoned & reduced core):::box
R3 -->|edit| R4
R4 -->|annotate| R5(fa:fa-cubes fa:fa-tags fa:fa-cc annotated,reasoned,reduced core):::box
R5 -->|convert| R6(fa:fa-star-o obo version):::box
R5 -->|convert| R7(fa:fa-star owl version):::box
classDef box font-family:avenir,font-size:14px,fill:#2a9fc9,stroke:#222,color:#fff,stroke-width:1px
</div>
<!--
[](https://mermaid-js.github.io/mermaid-live-editor/#/edit/eyJjb2RlIjoiZ3JhcGggVERcbiAgSTEoZmE6ZmEtdW5pdmVyc2l0eSBvbnRvbG9neSBzb3VyY2UgMSkgLS0-fGV4dHJhY3R8IE0xKGZhOmZhLWN1YmUgb250b2xvZ3kgbW9kdWxlIDEpXG4gIEkyKGZhOmZhLXVuaXZlcnNpdHkgb250b2xvZ3kgc291cmNlIDIpIC0tPnxleHRyYWN0fCBNMihmYTpmYS1jdWJlIG9udG9sb2d5IG1vZHVsZSAyKVxuICBJMyhmYTpmYS11bml2ZXJzaXR5IG9udG9sb2d5IHNvdXJjZSAzKSAtLT58ZXh0cmFjdHwgTTMoZmE6ZmEtY3ViZSBvbnRvbG9neSBtb2R1bGUgMylcbiAgTTEgLS0-IHxtZXJnZXwgUjEoZmE6ZmEtY3ViZXMgY29yZSlcbiAgTTIgLS0-IHxtZXJnZXwgUjEoZmE6ZmEtY3ViZXMgY29yZSlcbiAgTTMgLS0-IHxtZXJnZXwgUjEoZmE6ZmEtY3ViZXMgY29yZSlcbiAgUjEoZmE6ZmEtY3ViZXMgY29yZSkgLS0-IHxtYXRlcmlhbGl6ZXwgUjIoZmE6ZmEtY3ViZXMgcmVhc29uZWQgJiByZWR1Y2VkIGNvcmUpXG5cbiAgUjIoZmE6ZmEtY3ViZXMgcmVhc29uZWQgJiByZWR1Y2VkIGNvcmUpIC0tPnxyZXBvcnR8IFIzKGZhOmZhLXRhc2tzIHJlcG9ydClcbiAgUjIoZmE6ZmEtY3ViZXMgcmVhc29uZWQgJiByZWR1Y2VkIGNvcmUpIC0tPiB8ZWRpdHwgUjQoZmE6ZmEtY3ViZXMgcmVhc29uZWQgJiByZWR1Y2VkIGNvcmUpXG4gIFIzIC0tPnxlZGl0fCBSNFxuICBSNCAtLT58YW5ub3RhdGV8IFI1KGZhOmZhLWN1YmVzIGZhOmZhLXRhZ3MgZmE6ZmEtY2MgYW5ub3RhdGVkLHJlYXNvbmVkLHJlZHVjZWQgY29yZSlcbiAgUjUgLS0-fGNvbnZlcnR8IFI2KGZhOmZhLXN0YXItbyBvYm8gdmVyc2lvbilcbiAgUjUgLS0-fGNvbnZlcnR8IFI3KGZhOmZhLXN0YXIgb3dsIHZlcnNpb24pIiwibWVybWFpZCI6eyJ0aGVtZSI6Im5ldXRyYWwifX0) -->
## Capability & Maturity Table
<!-- TO DO -->
| Capability | Initial Maturity Level | Final Maturity Level |
| :------------- | :------------- | :------------- |
| Interoperability | minimal | repeatable |
----
## FAIRification Objectives, Inputs and Outputs
| Actions.Objectives.Tasks | Input | Output |
| :------------- | :------------- | :------------- |
| [ontology building](http://edamontology.org/operation_XXXX) | [tsv,OWL]|OWL|
## Table of Data Standards
| Data Formats | Terminologies | Models |
| :------------- | :------------- | :------------- |
| [OWL](https://fairsharing.org/FAIRsharing.atygwy) | | |
| [OBO](https://fairsharing.org/FAIRsharing.aa0eat) | | |
___
## Ingredients
| Tool Name | Tool Location | Tool function |
| :------------- | :------------- | :------------ |
| ROBOT | [http://robot.obolibrary.org/](http://robot.obolibrary.org/) | ontology management cli |
| Ontology development kit | [https://github.com/INCATools/ontology-development-kit](https://github.com/INCATools/ontology-development-kit) (comes with ROBOT included)| ontology management environment |
| Protégé/other ontology editor | [https://protege.stanford.edu/](https://protege.stanford.edu/) | ontology editor |
| SPARQL | [https://www.w3.org/TR/sparql11-query/](https://www.w3.org/TR/sparql11-query/) | ontology query language |
| ELK |[https://www.cs.ox.ac.uk/isg/tools/ELK/](https://www.cs.ox.ac.uk/isg/tools/ELK/)|ontology reasoner|
|Hermit|[http://www.hermit-reasoner.com/](http://www.hermit-reasoner.com/)|ontology reasoner|
## Step by step process
### Preliminary requirements
The development of `application ontology` requires joint contributions from `domain experts`, `use case owners` and `ontology developers`. The domain expert provides essential domain knowledge. The use case owners defines the `competency questions` of the application ontology. And the ontology developers are `IT specialists` working on the construction of the application ontology.
### Step 1: Define the goal of the application ontology
The development of an `application ontology` is driven by specific use cases and target datasets. The first step in application ontology development is to determine the subject and the aim of the application ontology.
In this recipe, we demonstrate the workflow of building an application ontology for patient metadata and patient sequencing data investigations. `Competency questions` of this ontology are provided below:
<!-- [here](https://github.com/FAIRplus/the-fair-cookbook/blob/ontology_robot_recipe/docs/content/recipes/ontology-robot/competency_questions.md). -->
| **Theme** | **Competency Questions** |
|:------------------------------------------------------|:--------------------------------------------------------------------------------------------------------------------------------------|
| **General Questions** | |
| | :heavy_plus_sign: How to identify relevant public domain ontologies suiting our needs? |
| | :heavy_plus_sign: How to establish an ontology covering all terms that are included in the actual data to be represented?
| | :heavy_plus_sign: How to remove terms from the resulting ontology that are not needed? |
| | :heavy_plus_sign: How to guarantee consistency of the final ontology? |
| | :heavy_plus_sign: How to identify differences in comparison to a previous version of the resulting ontology? |
| **Questions without specifying compounds or genes for the [example dataset](https://github.com/FAIRplus/the-fair-cookbook/blob/ontology_robot_recipe/docs/content/recipes/ontology-robot/ExternalStudiesKQ.xlsx)** | |
| | :heavy_plus_sign: Identify all data generated from tissues taken from patients suffering from a specific disease. |
| | :heavy_plus_sign: Identify all data generated from a specific tissues obtained from mouse models that are related to a specific disease. |
| | :heavy_plus_sign: Identify all data generated from lung tissue taken from patients suffering from a lung disease that is not related to oncology. |
| | :heavy_plus_sign: Identify all data generated from primary cells whose origin is the lung. |
| | :heavy_plus_sign: Identify all data generated from celllines whose origin is the lung. |
| **Questions including single genes or gene sets** | |
| | :heavy_plus_sign: What is the expression of PPARg / growths factors in lung tissue from IPF patients? |
| | :heavy_plus_sign: What is the expression of PPARg / growths factors in primary cells obtained from lung tissue from healthy subjects? |
| | :heavy_plus_sign: What is the expression of PPARg / growths factors in all available tissues obtained from healthy subjects? |
| **Questions including compound or treatment information** | |
| | :heavy_plus_sign: Identify all data generated from primary cells treated with a kinase inhibitor. |
| | :heavy_plus_sign: Identify all data from patients treated with a specific medication. |
| | :heavy_plus_sign: Identify all data generated from cells / celllines that have been treated with compounds targeting a member of a specific pathway. |
| | :heavy_plus_sign: What is the expression of PPARg in lung tissue upon treatment with a specific compound in patients suffering from a specific disease |
_Table 1_ is a snapshot of the example dataset. The complete patient metadata example dataset is [here](https://github.com/FAIRplus/the-fair-cookbook/blob/ontology_robot_recipe/docs/content/recipes/ontology-robot/ExternalStudiesKQ.xlsx).
| Study | source_id | sample_description | tissue | source_tissue | cell | cellline | disease | gender | species |
|----------------------|-----------|---------------------|--------|---------------|-----------------------|----------|-------------------------------|--------|--------------|
| GSE52463Nance2014 | EX08_001 | Lung - Normal | Lung | | | | Normal | male | Homo sapiens |
| GSE52463Nance2014 | EX08_015 | Lung - IPF | Lung | | | | Idiopathic Pulmonary Fibrosis | male | Homo sapiens |
| GSE116987Marcher2019 | EX101_1 | HSC CCl4-treated w0 | | | Hepatic Stellate Cell | | NA | | Mus musculus |
_Table 1: Patient metadata example_
### Step 2: Select terms from reference ontologies
#### Step 2.1 Select source ontologies
To build an `application ontology` that supports communication between different data resources, it is recommended to reuse existing terms from existing reference ontologies instead of creating new terms.
>:bulb: _Reference ontology:_ a semantic artifact with a canonical and comprehensive representation of the entities in a specific domain.
>
>:bulb: _Source ontology:_ An ontology to be integrated into the application ontology, usually a reference ontology.
When selecting reference ontologies as source ontologies, the reusability and sustainability of the source ontology need to be considered. Many ontologies in [the OBO foundry](http://www.obofoundry.org/) use the [CC-BY](https://creativecommons.org/licenses/by/2.0/) licence, allowing sharing and adaptation. Such ontologies can be directly used as source ontology. Reference ontologies with similar umbrella structures can be conveniently combined in the application ontology. The format and maintenance of the reference ontology shall also be considered.
Specific use cases and requirements from the target dataset also affect the choice of source ontologies. For use cases focusing on extracting data from heterogeneous datasets with complicated data types and data relationships, reference ontologies with rich term relationships are preferred. The interpretation of each term also depends on the context and requirements of the target dataset. For example, _"hypertension"_ can be either interpreted as a _phenotype_ and mapped to phenotype ontologies, or a _disease_ mapped to disease ontologies.
The example dataset includes metadata related to disease, species, cell lines, tissues and biological sex. _Table 2_ lists some reference ontologies available in corresponding domains. In this recipe, we selected _MONDO_ for disease domain, _UBERON_ for anatomy, _NCBItaxon_ for species taxonomy and _PATO_ for biological sex.
|Domain|Reference Ontologies|
|----|----|
|Disease| [Mondo Disease Ontology, MONDO](http://www.obofoundry.org/ontology/mondo.html)<br> [Disease Ontology, DIOD](https://disease-ontology.org/)<br> |
|Species|[NCBI organismal classification, NCBItaxon](http://www.obofoundry.org/ontology/ncbitaxon.html)|
|Cell line|[Cell Ontology, CL](http://www.obofoundry.org/ontology/cl.html)<br> [Cell Line Ontology, CLO](http://www.obofoundry.org/ontology/clo.html)|
|Tissue|[NCI Thesaurus OBO Edition, NCIT](http://www.obofoundry.org/ontology/ncit.html)<br> [Ontology for MIRNA Target, OMIT](http://www.obofoundry.org/ontology/omit.html)<br> [Uberon multi-species anatomy ontology, UBERON](http://www.obofoundry.org/ontology/uberon.html)|
|Biological Sex|[Phenotype And Trait Ontology, PATO](http://www.obofoundry.org/ontology/pato.html)|
_Table 2: Available reference ontologies in selected domains_
#### Step 2.2 Select seed ontology terms
> :bulb: _Seed ontology terms_: A set of entities extracted from reference ontologies for the application ontology.
>
This step identifies the seeds needed to perform the knowledge extraction from external sources, i.e., the set of entities to extract in order to be integrated on the application ontology. Ontology Developer can provide the tools to ease and to scale the identification of the seeds. Domain experts can identify the right seeds for a given application ontology.
Besides the fact that is always possible to manually identify the set of seeds needed for the performing of the concept extraction, to have a helper tool allows to run the task at scale. Following, an automatable approach based on using widely known life sciences annotators - [Zooma](https://www.ebi.ac.uk/spot/zooma/) and [NCBO Annotator](https://bioportal.bioontology.org/annotator) - are depicted.
<div class="mermaid">
graph LR
A(fa:fa-file-text-o Select seed terms):::box -->B1(fa:fa-user-o Manual selection):::box
A -->B2(fa:fa-database Selection with ZOOMA):::box
A -->B3(fa:fa-database Selection with NCBO Annotator):::box
A -->B4(fa:fa-cogs Selection with SPARQL):::box
classDef box font-family:avenir,font-size:14px,fill:#2a9fc9,stroke:#222,color:#fff,stroke-width:1px
</div>
_Figure 1: Ontology seed term selection approaches_
[ZOOMA](https://www.ebi.ac.uk/spot/zooma/) is a web service for discovering optimal ontology mappings, developed by the Samples, Phenotypes and Ontologies Team at the [European Bioinformatics Institute (EMBL-EBI)](https://www.ebi.ac.uk)
. It can be used to automatically annotate plain text about biological entities with ontology classes. Zooma allows to limit the sources used to perform the annotations. These sources are either curated datasources, or ontologies from the [Ontology Lookup Service (OLS)](https://www.ebi.ac.uk/ols/index). Zooma contains a linked data repository of annotation knowledge and highly annotated data that has been feeded with manually curated annotation derived from publicly available databases. Because the source data has been curated by hand, it is a rich source of knowledge that is optimised for this task of semantic markup of keywords, in contrast with text-mining approaches.
The [NCBO Annotator](https://bioportal.bioontology.org/annotator) is an ontology-based web service that annotates public datasets with biomedical ontology concepts based on their textual metadata. It can be used to automatically tag data with ontology concepts. These concepts come from the Unified Medical Language System (UMLS) Metathesaurus and the National Center for Biomedical Ontology (NCBO) BioPortal ontologies.
Both the `Zooma` and `NCBO Annotator service` provides a `web interface` and a `REST API` to identify the seed terms by annotation of free text. Two scripts able to automate the annotation of a set of free text terms are shown following.
#### Seed term extraction with ZOOMA
The following sample script uses the `Zooma web service` to get a list of seed terms - i.e., the URIs of ontology classes -. The service also states the level of confidence of every seed proposed.
```python3
#Python3
#zooma-annotator-script.py file
def get_annotations(propertyType, propertyValues, filters = ""):
"""
Get Zooma annotations for the values of a given property of a given type.
"""
import requests
annotations = []
no_annotations = []
for value in propertyValues:
ploads = {'propertyValue': value,
'propertyType': propertyType,
'filter': filters}
r = requests.get('http://www.ebi.ac.uk/spot/zooma/v2/api/services/annotate',
params=ploads)
import json
data = json.loads(r.text)
if len(data) == 0:
no_annotations.append(value)
for item in data:
annotations.append((f"{item['semanticTags'][0]} "
f"# {value}"
f"-Confidence:{item['confidence']}"))
return annotations, no_annotations
if __name__ == "__main__":
propertyType = 'gender'
propertyValues = ['male', 'female', 'unknown']
annotations, no_annotations = get_annotations(propertyType, propertyValues)
from pprint import pprint
pprint(annotations)
pprint(no_annotations)
```
Running the above script to get the seeds for the terms `male`, `female`, and `unknown` generates the following results:
```python3
['http://purl.obolibrary.org/obo/PATO_0000384 # male-Confidence:HIGH',
'http://purl.obolibrary.org/obo/PATO_0000383 # female-Confidence:HIGH',
'http://www.orpha.net/ORDO/Orphanet_288 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0003850 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0003952 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0009471 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0000203 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0003863 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0000616 # unknown-Confidence:MEDIUM',
'http://purl.obolibrary.org/obo/HP_0000952 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0003853 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_1001331 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0003769 # unknown-Confidence:MEDIUM',
'http://purl.obolibrary.org/obo/HP_0000132 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0000408 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0008549 # unknown-Confidence:MEDIUM',
'http://www.ebi.ac.uk/efo/EFO_0001642 # unknown-Confidence:MEDIUM']
[]
```
#### Seed term extraction with NCBO Annotator
The following sample script uses the NCBO Annotator web service to get a list of seed terms - i.e., the URIs of ontology classes -.
```python3
#Python3
#ncbo-annotator-script.py file
def get_annotations(propertyValues, ontologies = ''):
"""
Get NCBO Annotations for the values of a given property.
"""
import requests
annotations = []
no_annotations = []
for value in propertyValues:
ploads = {'apikey': '8b5b7825-538d-40e0-9e9e-5ab9274a9aeb',
'text': value,
'ontologies': ontologies,
'longest_only': 'true',
'exclude_numbers': 'false',
'whole_word_only': 'true',
'exclude_synonyms': 'false'}
r = requests.get('http://data.bioontology.org/annotator', params=ploads)
import json
data = json.loads(r.text)
if len(data) == 0:
no_annotations.append(value)
for item in data:
annotations.append(f"{item['annotatedClass']['@id']} # {value}")
return annotations, no_annotations
if __name__ == "__main__":
propertyType = 'gender'
propertyValues = ['male', 'female', 'unknown']
annotations, no_annotations = get_annotations(propertyType, propertyValues)
from pprint import pprint
pprint(annotations)
pprint(no_annotations)
```
Running the above script to get the seeds for the terms `male`, `female`, and `unknown` generates the following results:
```python3
['http://purl.obolibrary.org/obo/UBERON_0003101 # male',
'http://purl.obolibrary.org/obo/UBERON_0003100 # female']
['unknown']
```
#### Step2.2.3 Seed term extraction with SPARQL
Instead of manually maintaining a list of seed terms to generate a module, a term list can be generated on the fly using a `SPARQL query`. Here, we generate a subset of `UBERON` terms which have a crossreference to either `FMA (for human anatomy)` or `MA (for mouse anatomy)` terms, since our example datasets includes human, mouse and rat data.
```sql
PREFIX scdo: <http://scdontology.h3abionet.org/ontology/>
PREFIX obo: <http://purl.obolibrary.org/obo/>
PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>
PREFIX oboInOwl: <http://www.geneontology.org/formats/oboInOwl#>
PREFIX owl: <http://www.w3.org/2002/07/owl#>
SELECT ?uri WHERE {
{
{
?uri oboInOwl:hasDbXref ?xref .
}
UNION
{
?axiom a owl:Axiom;
owl:annotatedSource ?uri;
oboInOwl:hasDbXref ?xref .
}
}
?uri a owl:Class
FILTER isLiteral(?xref)
FILTER regex( ?xref, "^FMA|^MA:", "i")
}
```
### Step 3: Extract ontology modules from source ontologies
Module extractions from ontologies can be run manually and in an ad hoc fashion. We would however recommend to collect all steps together into a script or `Makefile` to avoid missing steps. `ROBOT` steps can in theory be chained together into single large commands. Practical experience however teaches that this can have unexpected consequences as well as making debugging difficult in the event of an issue. It is therefore advisable to split extractions and merges out into individual steps with intermediate artefacts which can be deleted at the end of the process chain.
#### Step 3.1 Get source ontology files
We recommend starting each (re)build of the application ontology with the latest versions of the source ontologies unless there is a good reason not to update to the latest version. Ideally, this should be done automatically, for example through a shell script that CURLs all ontologies from their source URIs, e.g.
```shell
curl -L http://purl.obolibrary.org/obo/uberon.owl > uberon.owl
```
#### Step 3.2 Choose ontology extraction algorithms
`ROBOT` supports two types of ontology module extraction, `Syntactic Locality Module Extractor` (SLME) and `Minimum Information to Reference an External Ontology Term` (MIREOT). `SLME` extractions can be further subdivided into TOP (top module), `BOT` (bottom module) and `STAR` (fixpoint-nested module). For full details of what these different options entail, see http://robot.obolibrary.org/extract.html. We recommend the use of BOT for comprehensive modules that preserve the links between ontology classes and the use of `MIREOT` if relationships apart from parent-child ones are less important.
#### Step 3.3 Extract modules using seed terms
Using `robot` tool provided by the `OBO foundry`, the creation of a module can be done with one command:
```bash
robot extract --method <some_selection> \
--input <some_input.owl> \
--term-file <list_of_classes_of_interest_in_ontology.txt> \
--intermediates <choose_from_option> \
--output ./ontology_modules/extracted_module.owl
```
The `robot` extract command takes several arguments:
* *method*: `ROBOT` uses 4 different algorithms to generate a module. `TOP`, `BOT`, `STAR` (all from the SLME method), and `MIREOT`. The first two will create a module below or above the seed classes (the classes of interest in the target ontology) respectively. The `STAR` method creates a module by pulling all the properties and axioms of the seed classes but nothing else. `MIREOT` uses a different methods and offers some more options, in particular when it comes to how many levels up or down (parent and children) are needed.
* *input*: this argument is to specify the target ontology you want to extract a module from. It can be the original artefact or a filtered version of it.
* *imports*: this argument allows to specific whether or not to include imported ontologies. Note that the default is to do so using the value `include`. Choose `exclude` otherwise.
* *term-file*: the text file holding the list of classes of interested identified by their iri (e.g. http://purl.obolibrary.org/obo/UBERON_0001235 # adrenal cortex)
* *intermediates*: specific to the `MIREOT` method, it allows to let the algorithm know how much or how little to extract. It has 3 levels (`all`,`minimal`, `none`).
* *output*: the path to the owl file holding the results of the module extraction
* *copy-ontology-annotations*: a boolean value true/false to pull or not any class annotation from the parent ontology. default is `false`
* *sources*: optional, a pointer to a file mapping
* *annotate-with-source*: a boolean value true/false. default is `false`
The above query, saved under `select_anatomy_subset.sparql` can be used to generate a dynamic seed list, then do a `BOT` extraction:
```shell
robot query --input uberon.owl --query select_anatomy_subset.sparql uberon_seed_list.txt
robot extract --method BOT --input uberon.owl --term-file uberon_seed_list.txt -o uberon_subset.owl
```
#### Step 3.4 Assess extracted modules
The extracted ontology module should include all seed terms and represent the term relationships correctly. It should also preserve the correct hierarchical structure of the source ontology and have consistent granularity.
### Step 4: Build the upper level umbrella ontology
The umbrella ontology is the root structure of the ontology. Building the umbrella ontology aims to model the main entity of the use case and its relationships with the ontology classes extracted on the previous step. The main identity of the ontology and relationships with extracted modules can be identified by domain experts, or specified by the use case owner.
`ROBOT` provides a template-driven ontology term generation system. A `ROBOT template` is a `tab-separated values` (TSV) or `comma-separated values` (CSV) file that depicts a set of patterns to build ontology entities - i.e., classes, properties, and individuals -. These templates can be used for modular ontology development.
After the _Domain Expert_ and the _Use Case/Scenario Owner_ specify the main entity of the use case and its relationships with remaining ontology entities, the _Ontology Developer_ generates the `ROBOT` templates depicting the set of patterns aimed to build the _umbrella_ ontology.
A `ROBOT` command using a template to build an ontology is shown below:
```bash
robot template --template template.csv \
--prefix "r4: https://fairplus-project.eu/ontologies/R4/" \
--ontology-iri "https://fairplus-project.eu/ontologies/R4/" \
--output ./templates/umbrella.owl
```
And a template sample is presented following:
| ID | Label | Entity Type | Equivalent Axioms | Characteristic | Domain | Range |
| -------- | -------- | -------- | -------- |-------- | -------- | -------- |
| ID | LABEL | TYPE | EC % | CHARACTERISTIC | DOMAIN | RANGE |
|ex:cl_2 | Class_2 | class |-|-|-|-|
|ex:cl_3 | Class_3 | class |ex:cl_2|-|-|-|
|ex:op_1 |Prop_1| object property|-|Class_2|Class_3| |
|ex:dp_1 |Prop_2| data property|-|functional|Class_2|xsd:string|
>:warning: **_Tip:_** The generated ontology can be visualized by using the Protege tool or a [local deployment of OLS](https://hackmd.io/@FAIRcookbook/H1axYY3wU). The `OLS local deployment` option is recommended by this recipe, given that `Protege` crash when loading medium or large size ontologies.
### Step 5: Merge ontology modules and umbrella ontology
Merging the ontology modules previuosly extracted and the umbrella ontology locally built produces a first draft of the application ontology.
The `merge` ROBOT command allows to combines two or more separate input ontology modules into a single ontology. Following, the `ROBOT` command merging the umbrella ontology and the modules is shown:
```bash
java -jar robot.jar merge \
--input ./ontology_modules/iao_mireot_robot_module_1.owl \
--input ./ontology_modules/obi_mireot_robot_module_2.owl \
--input ./ontology_modules/duo_mireot_robot_module_3.owl \
--input ./templates/umbrella.owl \
--output ./results/r4_app_ontology.owl
```
### Step 6: Post-merge modifications
#### Step 6.1: Materialize and Reasoning
Commands below infer superclasses and superclasses and reduce duplicated axioms merged term:
```bash
robot materialize \
--reasoner ELK \
--input merged_owl \
reduce \
--output materialized.owl
```
The ontology materialization uses `OWL reasoners`. `ROBOT` provides [several ontology reasoners](http://robot.obolibrary.org/reason).
It is also possible to identify issues in the ontology with some `quality control SPARQL queries`.
```bash
robot report --input edit.owl --output report.tsv
```
#### Step 6.2: Annotate
Ontology annotation adds metadata to the owl file. It is recommended to provide `ontology IRIs`, `version IRIs`, `ontology title`, `descriptions` and `license` to support future usage and management.
The annotation can be added either line-by-line or provided in a turtile(.ttl) file.
```bash
#ANNOTATE
robot annotate --input materialized.owl \
--ontology-iri "https://github.com/ontodev/robot/examples/annotated.owl" \
--version-iri "https://github.com/ontodev/robot/examples/annotated-1.owl" \
--annotation rdfs:comment "Comment" \
--annotation rdfs:label "Label" \
--annotation-file annotations.ttl \
--output results/annotated.owl
```
Below is an example annotation file.
```bash
@prefix rdf: <http://www.w3.org/1999/02/22-rdf-syntax-ns#> .
@prefix rdfs: <http://www.w3.org/2000/01/rdf-schema#> .
@prefix owl: <http://www.w3.org/2002/07/owl#> .
@prefix example: <https://github.com/ontodev/robot/examples/> .
@prefix dcterms: <http://purl.org/dc/terms/> .
example:annotated.owl
rdf:type owl:Ontology ;
rdfs:comment "Comment from annotations.ttl file." .
dcterms:title "Example title"
dcterms:license <http://creativecommons.org/publicdomain/zero/1.0/>
owl:versionIRI <https://github.com/ontodev/robot/examples/annotated-1.owl>
```
#### Step 6.3: Convert
Besides `OWL format`, the `Open Biomedical Ontology (OBO) format` is also widely used in life science related ontologies. It is possible to convert an `.owl` file to `.obo` file using:
```bash
#CONVERT:
robot convert \
--input annotated.owl\
--format obo \
--output results/annotated.obo
```
### Step 7: Assess coverage of the ontology scope
The final step of the ontology construction is to assess coverage of the ontology scope by verifying if it is able to answer the competency questions predefined. The competency questions can be implemented as a set of `SPARQL queries` and run against the developed ontology to check if the answers/results are aligned with the scope of the ontology. The `use case owner` and o`ntology developer` can also collaborate on the assessment of the competency questions answers/results.
`ROBOT` provides the `query` command to perform `SPARQL queries` against an ontology to verify and validate the ontology.
The query command runs SPARQL `ASK`, `SELECT`, and `CONSTRUCT` queries by using the `--query` option with two arguments: `a query file` and `an output file`. Instead of specifying one or more pairs (query file, output file), it is also possible to specify a single `--output-dir` and use the `--queries` option to provide one or more queries of any type. Each output file will be written to the output directory with the same base name as the query file that produced it. An pattern example of this command is shown following.
```bash
robot query --input <input_ontology_file> \
--queries <query_file> \
--output-dir <path_to_rsults> results/
```
---
## Conclusions:
> Creation an application ontology and semantic model to support knowledge discovery is an important process in the data management life cycle. This more advanced recipe has identified and described all the different steps that one needs to consider to build such a resource. While this is important, one should bear in mind the costs associated with maintaining those artefacts and keeping them up to date. It is therefore also critical to understand the benefits of contributing to existing open efforts.
> #### What should I read next?
> * [How to establish a minimal metadata profile?]()
> * [How to submit/request terms to an ontology ?]()
___
## References
- R.C. Jackson, J.P. Balhoff, E. Douglass, N.L. Harris, C.J. Mungall, and J.A. Overton. [_ROBOT: A tool for automating ontology workflows_](https://link.springer.com/epdf/10.1186/s12859-019-3002-3?author_access_token=bB8BLjFWrdh42vR6DjT-nG_BpE1tBhCbnbw3BuzI2RPCZ2BK7EeexaCNYfT-cCz8Q_mrZomT2_svoQf12CW661Sagzw6JGF9DhJq3Q3fTPdMGFMtais7MRgx8-kDhp6uC9g2qcVh5FumTsveV22XVQ%3D%3D). BMC Bioinformatics, vol. 20, July 2019
- Arp, Robert, Barry Smith, and Andrew D. Spear. _Building ontologies with basic formal ontology_. Mit Press, 2015.
## Supplementary material
- [Example Dataset](https://github.com/FAIRplus/the-fair-cookbook/blob/ontology_robot_recipe/docs/content/recipes/ontology-robot/ExternalStudiesKQ.xlsx)
- [Competency questions](https://github.com/FAIRplus/the-fair-cookbook/blob/ontology_robot_recipe/docs/content/recipes/ontology-robot/competency_questions.md)
- [Scripts and intermediate artifacts](https://github.com/FAIRplus/the-fair-cookbook/tree/ontology_robot_recipe/docs/content/recipes/ontology-robot)
## Authors
| Name | Affiliation | orcid | CrediT role |
| :------------- | :------------- | :------------- |:------------- |
|Danielle Welter | LCSB, University of Luxembourg| [0000-0003-1058-2668](https://orcid.org/0000-0003-1058-2668) | Writing - Original Draft, Code |
|Philippe Rocca-Serra| UOXF|[0000-0001-9853-5668](https://orcid.org/orcid.org/0000-0001-9853-5668)| Writing - Review, Code|
|Fuqi Xu|EMBL-EBI|[0000-0002-5923-3859](https://orcid.org/orcid.org/0000-XXXX-XXXX-XXXX)|Writing - Draft, Review|
|Emiliano Reynares| Boehringer Ingelheim|[0000-0002-5109-3716](https://orcid.org/0000-0002-5109-3716)|Writing - Review, Code |
|Karsten Quast|Boehringer Ingelheim|[0000-0003-3922-5701](https://orcid.org/0000-0003-3922-5701)| Writing|
___
## License:
<a href="https://creativecommons.org/licenses/by/4.0/"><img src="https://mirrors.creativecommons.org/presskit/buttons/80x15/png/by-sa.png" height="20"/></a>
 Sign in with Wallet
Sign in with Wallet

