---
tags: teaching, hpc0
---
# 2022-03 <br> HPC0: Introduction to Linux
Welcome to the hack pad for HPC0 course from Research Computing at the University of Leeds!
You can edit this document using [Markdown syntax](https://guides.github.com/features/mastering-markdown/).
## Contents
1. [Links to resource](#Links-to-resources)
2. [Further reading](#Further-reading)
3. [Agenda](#Agenda)
4. [Pre workshop prep](#Pre-workshop-prep)
4.1. [Windows Users](#For-Windows-Users)
4.2. [MacOS/Linux Users](#For-MacLinux-Users)
6. [What's your name and where do you come from?](#What’s-your-name-and-where-do-you-come-from)
## Links to resources
- **Contact Research Computing** - https://bit.ly/arc-help
- **Request HPC account** - https://leeds.service-now.com/it?id=sc_cat_item&sys_id=4c002dd70f235f00a82247ece1050ebc
- **Slides for today** - https://bit.ly/hpc0linux
- **Exercises for today** - https://drive.google.com/file/d/1dV8fMS_n6GOFZO_rmFfUBwnuBFGj6C58/view?usp=sharing
## Further reading
Linux crib sheet that covers lots of idea from today and beyond - https://drive.google.com/file/d/0B4hIpRJzq8DPVG5xdEJWcGlRTkU/view?usp=sharing
## Agenda
| Time | Agenda |
| -------- | ------------------------------- |
| 0900 | Introduction, connecting to ARC |
| 0950 | Break |
| 1000 | Navigating the shell |
| 1050 | Break and Exercise 1 |
| 1100 | Data transformation in the shell|
| 1150 | Wrap up and questions |
| 1200 | Finish |
## Pre workshop prep
***If you haven’t already request an account for the HPC via this link - https://leeds.service-now.com/it?id=sc_cat_item&sys_id=4c002dd70f235f00a82247ece1050ebc***
For Windows users please consult our documentation page and video at https://arcdocs.leeds.ac.uk/getting_started/logon.html#connecting-from-windows
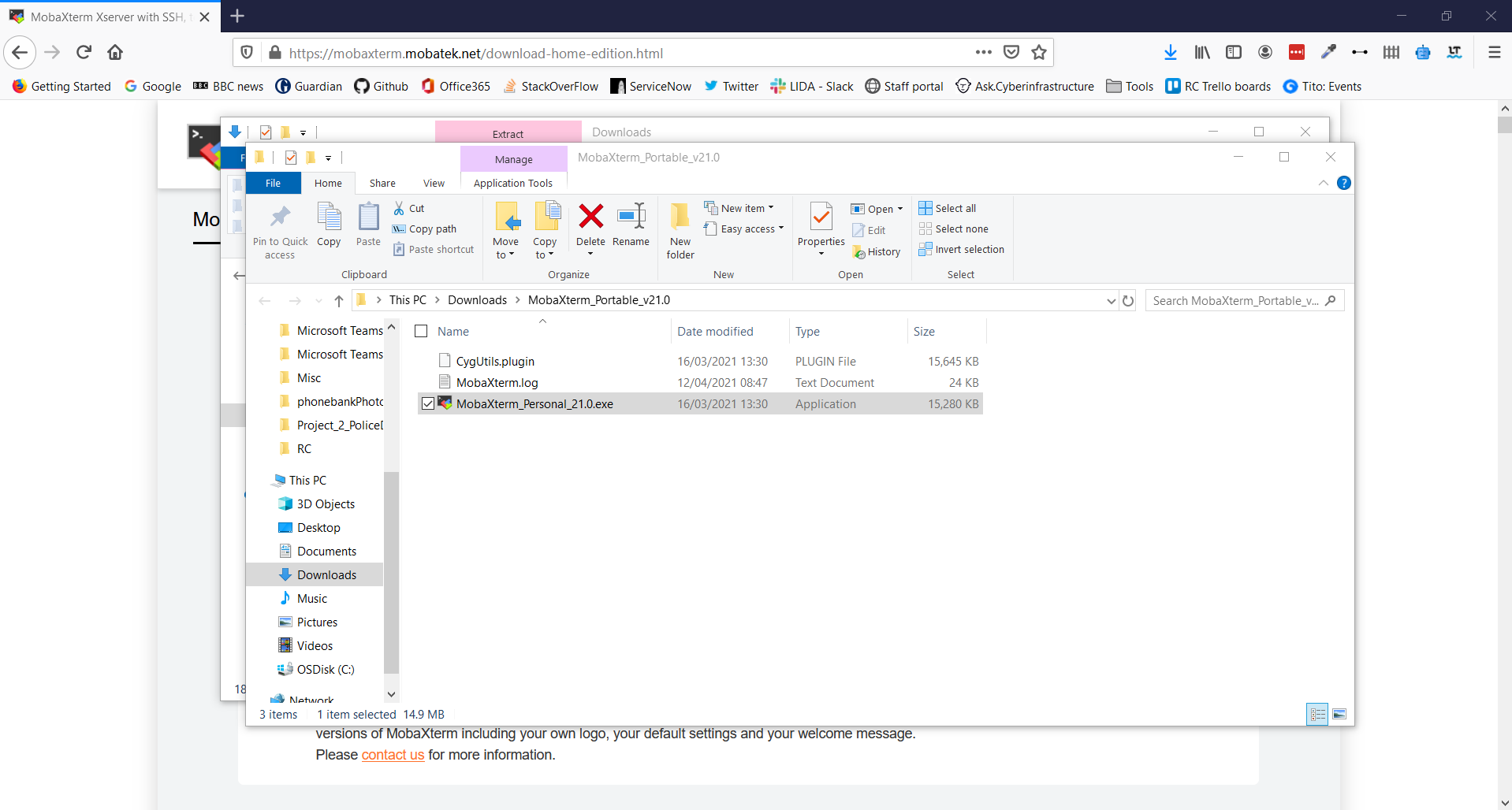
You are required to download the software tool MobaXTerm for this workshop.
1. Navigate using a web browser to https://mobaxterm.mobatek.net/
2. Select Download
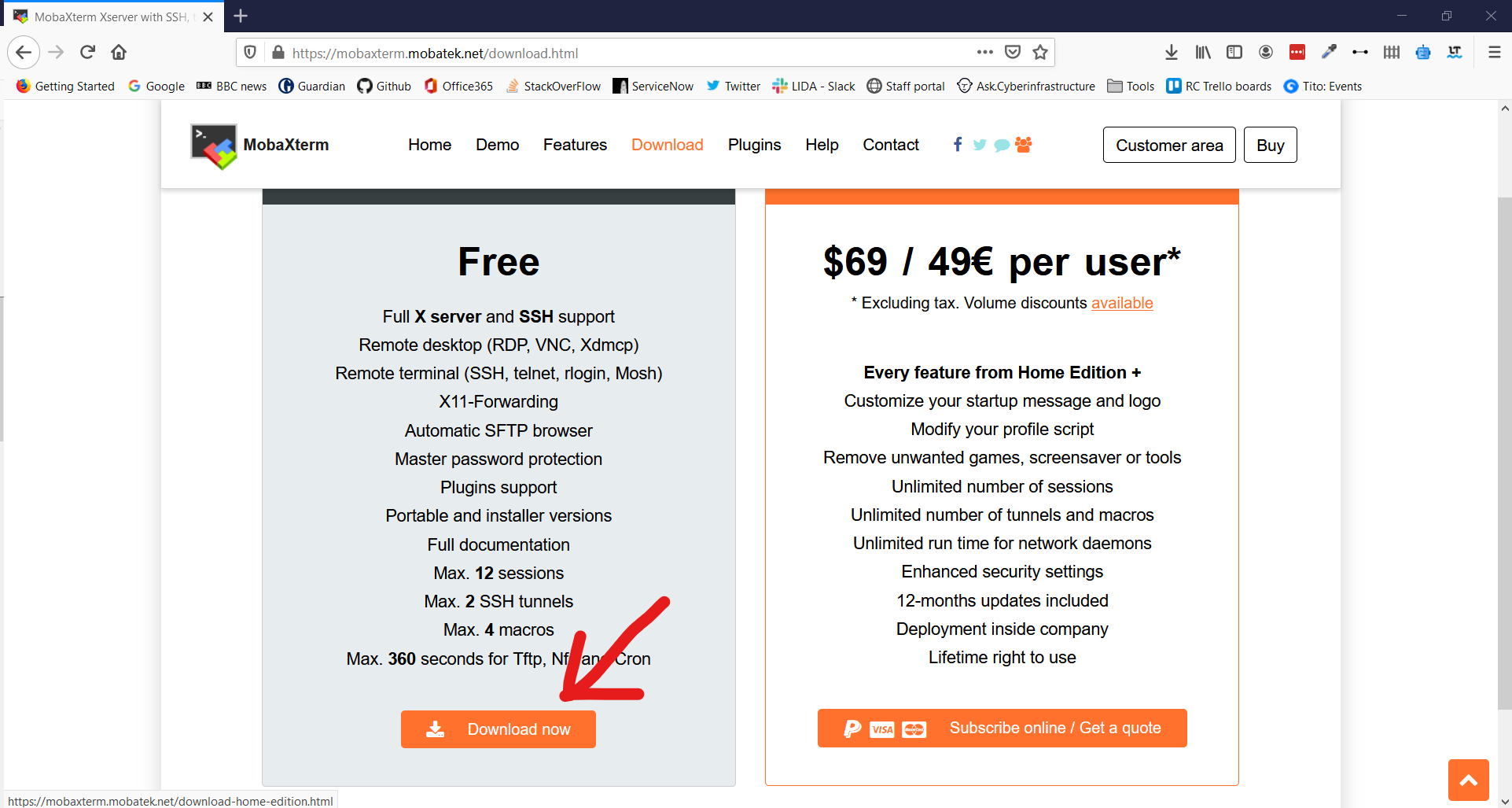
3. Click Download Now for the Home Edition
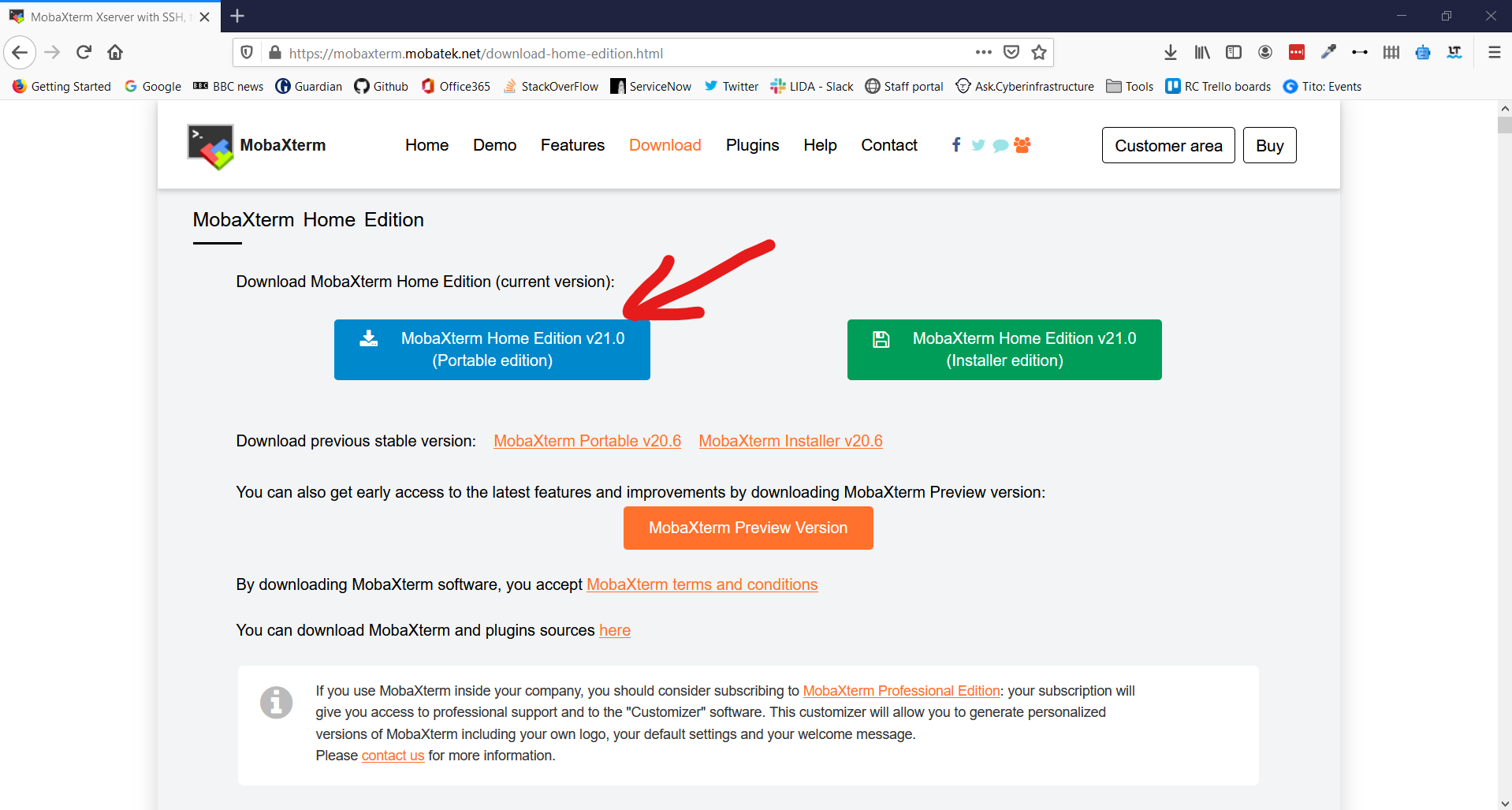
4. Select MobaXTerm Home Edition v21.0 (Portable edition)
5. This opens a download prompt for a .zip file. Select Save File and click OK
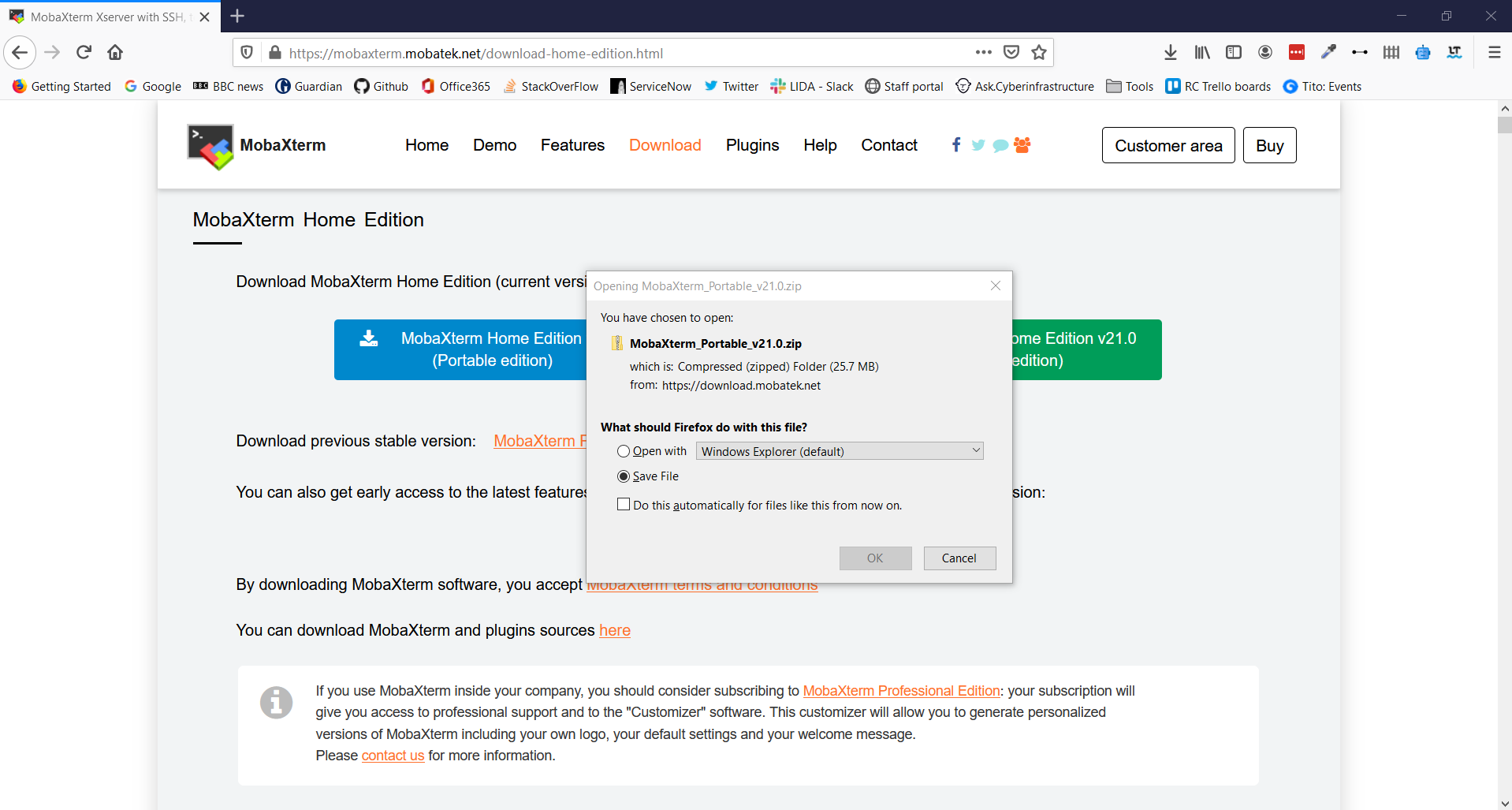
6. Go to your Download folder and find the .zip file you have just downloaded
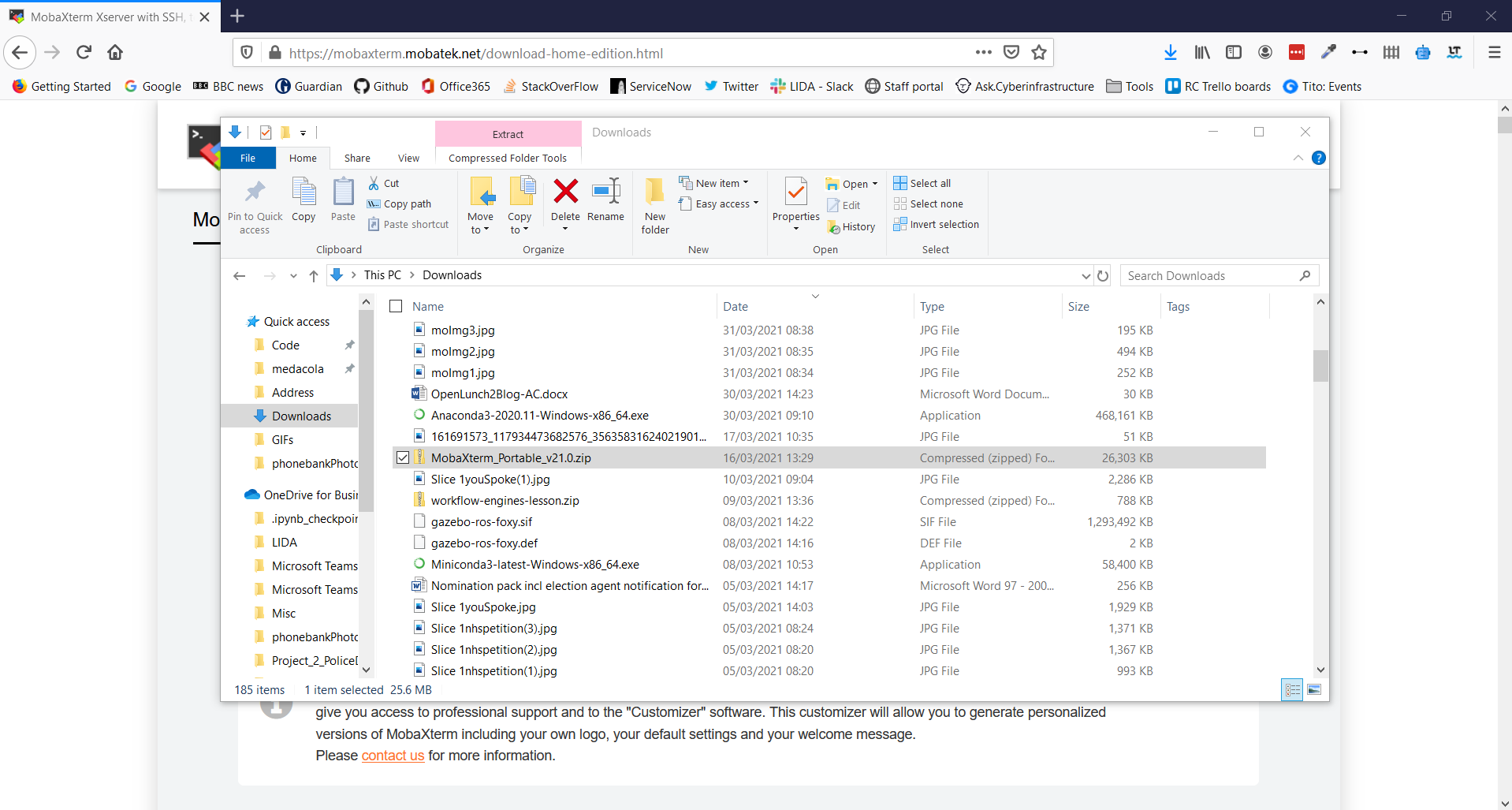
7. Click Extract in the Ribbon Bar and select Extract All
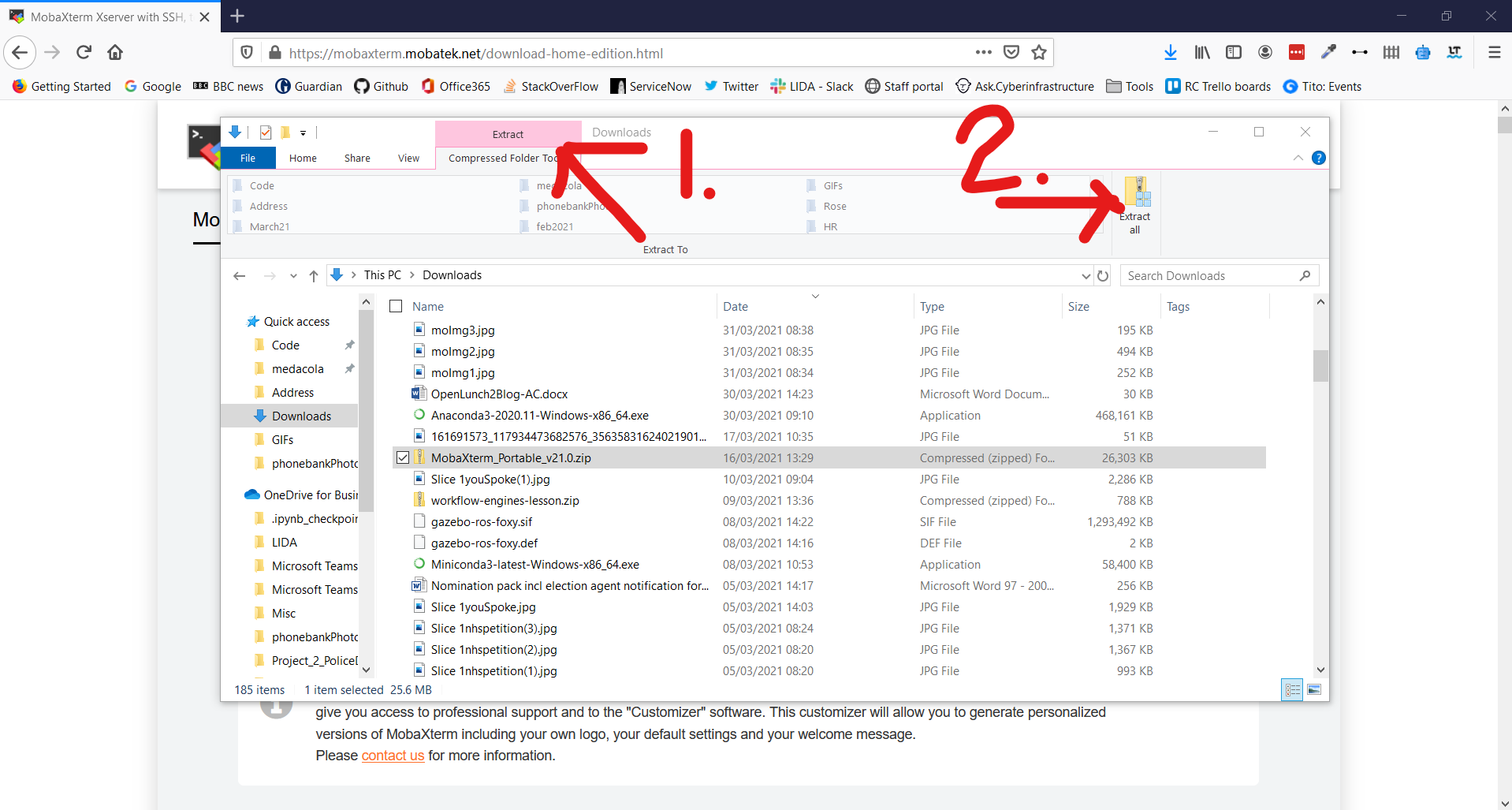
8. Using the Wizard window extract the folder at the suggested location
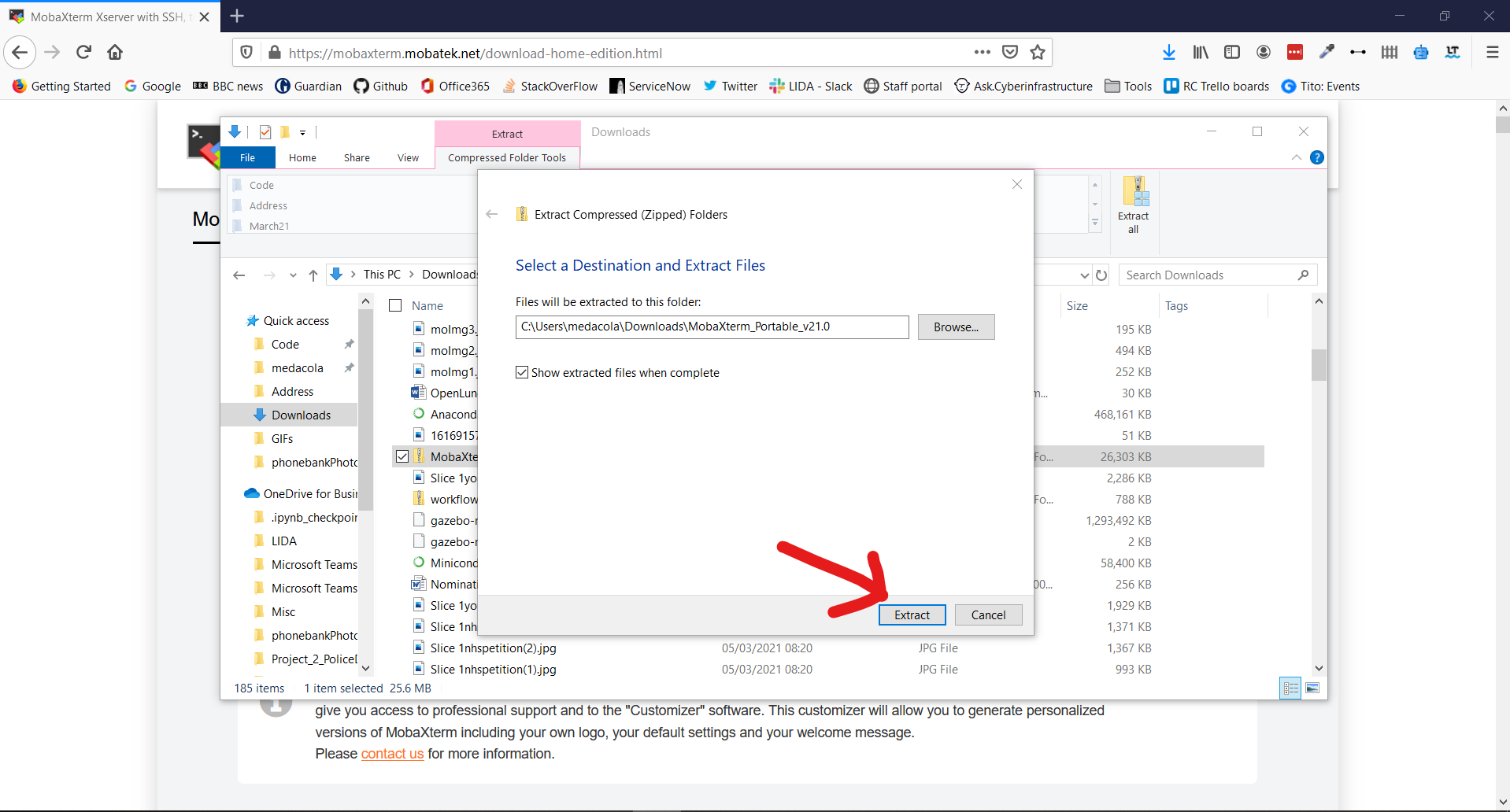
9. This should open the extracted folder immediately and allow you to double-click on the MobaXTerm_Personal_21.0 executable to start the application
**And you're all set for HPC0!🎉**
### For Mac/Linux Users:
**MacOS and Linux users do not need MobaXTerm** but can use your builtin Terminal application. You should follow the steps outlined in the bitesize video titled [“Connecting to ARC off-campus via Linux/MacOS”](https://arc.leeds.ac.uk/help/videos/) and read carefully the [documentation section](https://arcdocs.leeds.ac.uk/getting_started/logon.html#connecting-from-linux-macos-systems) on connecting from Linux and MacOS, especially the section about configuring SSH for off-campus connections.
Please attempt to read the documentation and watch the videos carefully and attempt the setup steps. If you have any problems don’t worry we will spend the start of the workshop making sure everyone is connected.
In order to connect to ARC when you're off campus you'll need to do some extra configuration so that your SSH connection goes via our `remote-access` server. The following steps outline how to setup this configuration:
1. Open a Terminal on your Linux/macOS machine
2. Create a directory called `.ssh` in your home directory (if one doesn't already exist)
```bash
$ mkdir ~/.ssh
```
3. Then open a text editor of your choice and create a file called `config` in your `.ssh` directory
```bash
# for instance use the simple nano text editor
$ nano ~/.ssh/config
```
4. Within this file include the following contents where `USERNAME` is replaced by your university username
```bash
Host *.leeds.ac.uk !remote-access.leeds.ac.uk
ProxyJump USERNAME@remote-access.leeds.ac.uk
User USERNAME
```
5. Save this file and your configuration is all set up!
## What's your name and where do you come from?
- Gamal Ahmed, PhD student (School of Earth and Environement)
- Raghda, PhD Research student, Medical school, dentistry, have no background in data science.
- Alex Coleman, Research Software Engineer, who likes linux, python and R. And has a background in data science and machine learning.
les, technician, Civil Eng
- Jessica Bryon, PhD Student, School of Medicine, RNA Sequencing.
- John Hodrien, Research Software Engineer within Research Computing (Linux,HPC,C/C++/Visualisation)
- Ollie Clark, Research Software Engineer. Web, databases and software engineering.
- Khushboo Gurung, PDRA, Climate modelling
- Luke Conibear, Research Software Engineer
- Andreas Kosteletos, PhD student looking at lncRNAs. Here today to learn about HPC
- Tom Dixon, Chemistry (Phd student). Testing optimsation algorithms on ML models
- James Macdonald, Research fellow working on new manufacturing processes in Chemical Engineering, intro to linux and hpc
- Sean, PGT at the School of Mechanical Engineering.
- Alex Scoffield, PhD Student (School of Geography). Examining how proglacial lakes have influenced glacier dynamics in the Himalayas using satellite imagery.
- Daniel Colson, PhD Student, School of Geography. Working with satellite imagery to explore regions of northern peatlands.
- Shradhanjali Sahu, PhD student
- Thiago Falcao, Phd Student, School of Earth and Environment, Numerical Modelling applied to Geomechanical Modelling.
- Nick Cooper, Medical Engineer in iMBE doing a PhD modelling replacement hip components.
- Jaimin Radia, Masters student studying chemical engineering
- Faza Bastarianto, PGR at Institute for Transport Studies
- Scott Grossman, research fellow in chemistry working on novel antibiotics. Hopefully going to be using HPC for some virtual screening and docking
- Charles Parton-Barr, taught postgraduate Physics student
- Kristina Bratkova, Data Scientist Intern, Leeds Institute for Data Analytics, interested about Linux command line
- I am Hanan Dreiwi a research vistor at the University at the school of mathematics. I would like to learn more about using ARC.
- Hannah Taylor, PhD student in Medicine and Health, interested in learning how to analyse data (RNAseq)
- Latifa, PhD student , cloud computing.
- Emily Caseley, postdoc in the Faculty of Biological Sciences, interested in learning how to run virtual screens for drug discovery.
- Khulood Alazwary, PhD student ,Electrical and Electronic Engineering.
-
- i am Adesola Olalekan, A visiting researcher in Prof Alex O' Neill group. My research is on USING BIOINFORMATICS tools to answer research questions on AMR from sequencing data.
- Deborah Olukan, PhD Student, Data Analytics and Society