# Comparative Analysis of ViT and ResNet Architectures for Disease Detection in Tomato Leaf Images
**Authors**
- Jolle Verhoog (j.t.verhoog@student.tudelft.nl)
- Petar Petrov (P.I.Petrov@student.tudelft.nl)
- Stan Marseille (s.r.marseille@student.tudelft.nl)
**Code:** https://github.com/petaripetrov/cv-tomato-leaf
*This blogpost was written as part of the Seminar Computer Vision by Deep Learning course at Delft University of Technology.*
## Introduction
Early and accurate identification of diseases in plants may increase farm yields and reduce pesticide usage, contributing to efficient use of agricultural resources and reduced environmental impact. In agriculture and floriculture, disease detection in plants is essential for productivity, ecological sustainability and economic stability [[15]](#15).
### Background
Research has demonstrated the potential for deep learning in detecting and classifying diseases in plants. For example, studies such as [[4]](#4) and [[5]](#5) suggest various diseases in plant species can be detected with high accuracy. In addition, algorithmic detection potentially allows for analyzing of vast amounts of data simultaneously and efficiently. As a result, algorithmic detection can offer accuracy and efficiency advantages over manual detection by humans [[6]](#6).
### Relevance
Developments in transformer based models have demonstrated significant potential for image recognition in terms of accuracy and efficiency in comparison to architectures such as convolutional neural networks (CNN), as shown by [[1]](#1), which introduced the ViT architecture. ViT models split an image into a set number of patches and treat the patches as individual tokens for the transformer, borrowing from modern Natural Language Processing practices. By doing this, ViTs are able to preserve the information in the image while keeping their parameter size low, and thanks to self-attention they achieve remarkably high performance [[10]](#10). More specifically for disease classifications, [[2]](#2) suggests ViT models may perform well when classifying x-ray images compared to CNNs. Given the performance of ViT models for identifying abnormalities, we investigate to what extent do ViTs generalize efficiently on visible diseases in tomato plants.
### Pretrained Models
The following state-of-the-art pretrained models were used: ResNet-50 [[7]](#7) and ViT-base-patch16-224-in21k [[12]](#12). ResNet-50 is a convolutional model that has 50 layers and addresses vanishing gradients through skip connections. With these skip connections the gradient bypasses any layer that ends up damaging the performance. This is the reason why ResNet-50 has impressive performance on, for instance, ImageNet [[13]](#13) when compared to similar models. ResNet has 26 million parameters and below is a schemtaic of the model is shown.

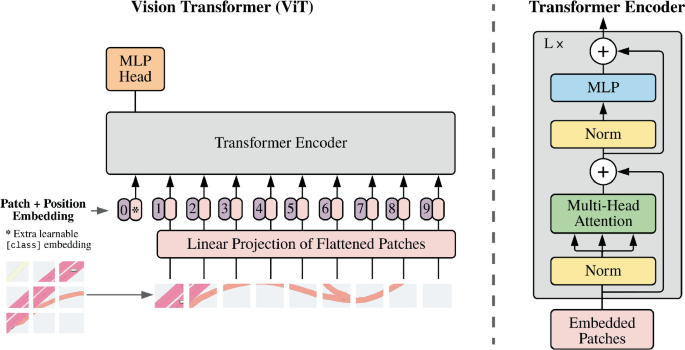
The transformer model we use, ViT-base-patch16-224-in21k, was introduced in [[13]](#13). This vision transformer divides 224-pixel square images into 16 patches of 16x16 pixels. These patches are positionally embedded and passed through 12 encoder layers. This model does not include any finetuned heads, allowing us to add our own specific to our image classification task. This transformer model has 86 million parameters and a visualisation is shown below.
<a id="related-works"></a>
## Related Work
Different studies have investigated the effectiveness of deep learning, CNNs and ViTs in disease classification problems, and disease classification in tomato plants. This section summarizes the primary studies we consulted in designing our investigation.
Firstly, Ahmed et al. (2022) [[4]](#4) proposed ‘a lightweight approach’ deep learning approach for classifying diseases in tomato plants. PlantVillage data was preprocessed and enhanced using CLAHE (Contrast Limited Adaptive Histogram Equalization). A pre-trained MobilenetV2 architecture was used for feature extraction and training the model.
The study found the MobileNetV2 architecture achieved 99.30% mean accuracy on test sets. In addition, implementation of CLAHE demonstrated a positive effect on test accuracy, improving a baseline accuracy of 97.27% to 97.71%. At the same time, the model was small in size (9.6 MB) and complexity (4.87M flops).
Ahmed et al. suggest an effective solution for classifying diseases in tomato plants using the MobileNetV2, a CNN. Their experiments include results of computational complexity and model size. As such, this study serves as a benchmark for this project, suggesting lightweight, pre trained CNNs are efficient and yield strong results. In addition, the authors provide insights in their approach for implementation and measurements of results, such as the implementation of CLAHE and data augmentation, and their measurements of computational complexity.
Secondly, Uparkar et al. (2023) [[2]](#2) compared the performance of ViTs to CNNs for x-ray lung-disease classification problems. The authors used ViT models and compared them with a hybrid CNN based VDSNet architecture, both pretrained on the ImageNet dataset. Again, feature extraction was used, using the VGG-16 model.
Uparker et al. found that the ViT models achieved similar or slightly better results compared to the VDSNet model. For example, ViT-Huge achieved 70.24% accuracy compared to 69.86% for VDSNet in similar conditions. They concluded that ViTs benefit from smaller patch sizes, deeper layers, and pre-training.
This study suggests performance increases for disease classification in images using ViTs over CNNs may be possible. This supports the relevance of our study. In addition, the authors provide a detailed approach for the implementations, experiments, and evaluation metrics. The study functions as a significant guideline for our approach.
## Hypothesis
Through experimental comparison, we primarily investigate to what extent ViT models may offer performance benefits compared to CNN-based ones for tomato disease detection. We also compare the performance of pretrained and non-pretrained ViTs and CNNs. Specifically, we investigate:
- Which architecture performs better for classifying tomato diseases from leaf images?
- To what extent does ImageNet-pretraining ResNet and ViT affect classification accuracy in classifying tomato diseases from leaf images, compared to training from scratch?
- To what extent does ImageNet-pretraining ResNet and ViT affect the training and inference time, compared to training from scratch?
We hypothesize that untrained and pretrained ViTs will achieve similar or higher accuracies when compared to their respective ResNet counterparts. We also hypothesize that training and inference times for ViTs will be higher than that for the ResNet architectures.
As described in the ['Related Works'](#related-works) section, [[2]](#2) found that ViTs achieve similar or slightly higher accuracy scores compared to CNNs architectures. ViTs use the self-attention mechanism on the trainable patches. Maurício et al. (2023) [[10]](#10) suggest this enables ViTs to capture global image information better, as information is accessible 'from the lowest to the highest layer'.
We expect global context to be relevant in tomato plant disease data. The disease may affect different parts of a plant, spreading the evidence across an image. As result classification of a disease may require a more global understanding of different parts of the data. At the same time, when global context is less relevant, this advantage may be limited.
In addition, [[3]](#3) and [[10]](#10) conclude that, while ViTs benefit from smaller patch sizes and more layers, this also increases the number of trainable parameters. As result training and inference times may increase as well.
## Experiments
Our study compares performance results of two types of architectures on the same dataset. This section describes the approach and steps to ensure the validity of our experiments.
### Experimental Setup
We implemented our experiments through a Jupyter notebook. We did all of the development and testing on Google Collab; however, due to GPU access limitations, we ran the experiments locally on an Nvidia Quadro P1000 with 512 Cuda cores with 4 Gb dedicated memory, 8 Gb system memory and an Intel Core i7-8750H CPU.
### Data Preparation
We use the "Tomato Leaf Disease Image" dataset hosted on HuggingFace [[9]](#9), which includes 18160 images representing ten classes. All of the images in the dataset are of size 256 by 256 and contain one tomato leaf with only one class. Originally, this dataset was intended for Text-to-Image tasks, so labels were of the format "This is an image of a tomato leaf with X" where X is the name of a disease. We simplified the labels to only include the disease shown in a given image and created an ID mapping as is standard for image classification tasks. Afterward, we split the dataset into subsets containing 80% of the original subsets for training, 10% for testing, and 10% for validation.
Further, we apply the individual preprocessing pipelines required by ViT and ResNet, which resize the images to 224 by 224 and normalize across the RGB channels. We present some examples from the dataset.
<div style="display: flex; align-items: center; justify-content: space-between;">
<div style="text-align: center;">
<img src="https://hackmd.io/_uploads/B1QU3RKS0.jpg" alt="bacterial spot" style="width: 100%;">
<p>Spider mite</p>
</div>
<div style="text-align: center;">
<img src="https://hackmd.io/_uploads/B1XL2RYS0.jpg" alt="spider mite" style="width: 100%;">
<p>Bacterial spot</p>
</div>
</div>
### Models
We import the models described in Section 1.3. In particular, we import the versions of ViT [[8]](#8) and ResNet-50 [[11]](#11) pre-trained on the widely used ImageNet-1k dataset. We additionally configure untrained versions of the models so we can measure the effects pre-training has on performance. We base our configurations on the ones defined by the pre-trained model; this is to say, the only difference between the un-trained and the pre-trained models is that the pre-trained models use publicly available checkpoints hosted on HuggingFace.
### Model Training and Tuning
To train and fine-tune our models, we set up a list with the necessary objects, such as the individual models, training parameters, prepared dataset, and the given model's preprocessing pipeline. We then iterate over the list and create a relevant Trainer class, an abstraction provided by HuggingFace that handles the training for us. After training the models, we save a snapshot of the parameters alongside the training metrics and evaluate the model on the test fold to measure the performance against unseen data. The models were trained with the following hyperparameters:
> **Table 1: Training Parameters**\
| Name | ViT | ResNet |
| -------- | -------- | -------- |
| Epochs | 3 | 3 |
| Batch size | 32 | 32
| Seed | 42 | 42|
| Learning rate | $2 * 10^{-4}$ | $5 * 10^{-5}$ |
### Loss function
$$
-\sum_{i=1}^Ct_i\log(p_i)
$$
We employed cross-entropy loss, as seen above, implemented in the HuggingFace Trainer class. The loss value is determined by comparing the predicted probability distribution ‘p_i’ to the actual probability distribution ‘t_i’ for class ‘i’ out of all classes ‘C’, making it particularly effective for multi-class classification problems like ours.
### Evaluation Metrics
To compare the classification performance of our models, we measure how well they can classify unseen data. In particular, we measure the accuracy, which is the ratio between the total correct classifications for all classes over all of the classifications made. In this context, correct means that the model predicted the label asigned in the dataset. Additionally, we measure the training and inference durations for all four models. We use these to compare the classification efficiencies of the used models.
In pursuit of fairness, we conducted the training and evaluation on the same machine and fixed the Trainer class' random generator seed.
### Assumptions and Limitations
During our experiments, we experienced issues primarily stemming from the "transformers" library provided by HuggingFace. In particular, we noticed that using the default classification head attached by the ResNetForImageClassification class causes the model to experience vanishing gradients under some randomizer seeds. We found that overriding the linear layer in the classification head with a new torch layer fixes this problem. We believe this may be due to the difference in the number of classes between our dataset and ImageNet because we get an error warning us that the classifier's bias and weight tensor sizes of [1000] and [1000, 2048], respectively, do not match our initialized sizes of [10] and [10, 2048]. We did not observe this error when initializing the ViT models or with the empty ResNet model.
We have cause to believe that the ResNet classes provided by HuggingFace might not be as optimized as the ViT ones, because the model uses the ConvNeXt preprocessor class instead of a ResNet-specific one. While this is not necessarily a problem, and we observed that our model trains without significant issues, we see this as a sign that more work is needed to provide a better experience for ResNet users. HuggingFace also provides a method for lazy-loading dataset preprocessing operations with the goal of optimizing computation. However, when utilizing this feature alongside the ResNet preprocessor we encountered errors during training telling us that the model only received label data from the batches and no image data. We were able to resolve this issue by processing the entire dataset at once, instead of using lazy-loading, but we believe this is even more proof that there might be issues with the implementation of some classes provided by HuggingFace.
## Results
After training and evaluation of the models as described in the 'Experiments' section, the results show differences between the ResNet and ViT model. This section summarizes those differences.
### Training:
With batches of 32 samples, and three epochs, training involved 1,362 steps per model. In addition, validation losses and accuracy were evaluated every 100 steps. The 'Total Training Time' indicates the duration for completing the entire process, including the validation evaluations. In training with 3 epochs, 43,584 samples were evaluated. 'Training speed' was determined by dividing 43,584 by the total durations spend processing all training samples per model, not validation samples.
> **Table 2: Training Metrics**
| Model | Average Training speed <br> (samples / sec.) |Total Training Time <br> H:M:S |
|:---:|:---:|:--------:|
| ResNet (untrained) | 5.142 |2:21:16 |
| ResNet (pretrained)| 3.707 | 3:15:58 |
| ViT (untrained) | 2.647 |4:34:25 |
| ViT (pretrained)| 2.319 |5:13:12 |
In the training process, the ResNet models showed higher training speeds than the ViT models. Both the untrained ResNet and ViT trained faster than the respective pretrained models. The differences in training speeds are more significant for ResNet than ViT. Untrained ResNet trained 39% faster than pretrained, while untrained ViT trained 14% faster than pretrained.
### Evaluation
In determining accuracies, all models were evaluated on the 1,816 unseen samples. In addition, average inference times per model were determined via random sampling from the test set. For every model, 100 samples were randomly drawn, resulting in 4 sets of 100 in total. The 'Inference Speed' was determined by dividing 100 samples by the inference time.
> **Table 3: Evaluation Metrics** <a id="table-3"></a>
| Model | Test Accuracy |Inference Speed <br> (samples per sec.)|
|:-------------|:--------------:|:--------------:|
| ResNet (untrained) | 0.8651 |4.4880 |
| ResNet (pretrained)| 0.8216 |22.413 |
| ViT (untrained) | 0.9058 |13.996 |
| ViT (pretrained) | 0.9983 |20.490 |
As Table [3](#table-3) shows, both ViT models achieved higher accuracies than the ResNet models on our test set. Pretrained ViT achieved a higher accuracy than untrained ViT, 99.83% and 90.58% respectively. However, pretrained ResNet preformed worse than untrained, with respective 82.16% and 86.51% test accuracies.
The results suggest pretrained ResNet and ViT achieve higher inference speeds than their untrained counterparts. The inference speed difference between untrained and pretrained ResNet is significantly higher than the difference between the ViT models.
## Discussion of results:
First we will recap our three main research questions and then we will answer them below. Our reserach questions were:
- Which architecture performs better for classifying tomato diseases from leaf images?
- To what extent does ImageNet-pretraining ResNet and ViT affect classification accuracy in classifying tomato diseases from leaf images, compared to training from scratch?
- To what extent does ImageNet-pretraining ResNet and ViT affect the training and inference time, compared to training from scratch?
As seen in the results, both ViT models outperformed the ResNet models in terms of accuracy on this dataset. The pretrained ViT achieved the highest test accuracy on our data. This may indicate that this transformer network is more suitable for transfer learning on downstream tasks than the pretrained ResNet model. Furthermore, when using untrained models, the ViT also slightly outperformed the ResNet model. Given the relatively small amount of epochs, this might also be due to randomness and further research could point this out. The better performance of the ViT models might also be due to the data being more suitable for ViT, because of an inherent quality we did not account for.
Given these findings we conclude that the better architecture for our dataset is the ViT. If both models are pretrained on ImageNet, only the ViT reaches high accuracy while the accuracy of the ResNet model is worse than the untrained model. We think this might be due to the thousand classification heads that the pretrained ResNet model includes, which can't be overwritten easily. This also induces more complexity than training an empty model to fit 10 classifcation heads. It is also possible that, simply, ResNet requires more epochs to properly fine-tune a dataset; meaning that, ViTs, or transformer-based models in general, benefit more from fine-tuning.
Regarding our third research question, for both models the inference time per sample was significantly better when a pretrained model was used. We think this is due to the model parameters being optimized better or due to optimizations included by the orignal authors. We hypothesized that the ViT models will be slower, but it seems that only the pretrained ResNet model performs faster. The respective training time of the ViT models is comparitvely higher, which may be due to the added complexity of the transformer-based architectures. In general, it seems that, computational performance wise, the pretrained ResNet model performs the best, making it an appropriate choice in situations where performance is key, if trained for enough epochs.
In conslusion, we have shown that when using comparable convolutional and transformer models, the transformer model provide better accuracy than the convolutional ones for classification of tomato diseases in leaves. We think this may be due to the diseases affecting different parts of the leaf and the transformer being better at 'seeing' the whole picture. However, in performance critical scenarios, it might be best to use a CNN model.
## Work Distribution
Petar: Writing of Experiments and configuring pipeline for training of both models
Stan: Writing of the HL story line, Introduction, Related works, Hypothesis and Results. Running the pipeline
Jolle: Writing of the HL story line, Introduction, Discussion of results
## References
[comment]: <> (Use APA 7th for references!)
<a id="1">[1]</a> Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., Uszkoreit, J., & Houlsby, N. (2020, October 22). An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv.org. https://arxiv.org/abs/2010.11929
<a id="2">[2]</a> Uparkar, O., Bharti, J., Pateriya, R., Gupta, R. K., & Sharma, A. (2023). Vision Transformer outperforms deep convolutional neural network-based model in classifying x-ray images. Procedia Computer Science, 218, 2338–2349. https://doi.org/10.1016/j.procs.2023.01.209
<!-- <a id="3">[3]</a> Hettiarachchi, H. (2023, August 12). what is Vision Transformers | Medium. Medium. https://medium.com/@hansahettiarachchi/unveiling-vision-transformers-revolutionizing-computer-vision-beyond-convolution-c410110ef061#:~:text=ViT%20employs%20self%2Dattention%20mechanisms,layers%20for%20coarse%20global%20information.
<a id="4">[4]</a> Joshi, S. (2022, October 19). Everything you need to know about : Inductive bias - Sharad Joshi - Medium. Medium. https://medium.com/@sharadjoshi/everything-you-need-to-know-about-inductive-bias-4850b77048f2#:~:text=This%20time%20the%20bias%20is,another%20inductive%20bias%20for%20CNNs. -->
<a id="3">[3]</a> Pu, Q., Xi, Z., Yin, S., Zhao, Z., & Zhao, L. (2024). Advantages of transformer and its application for medical image segmentation: a survey. BioMedical Engineering Online, 23(1). https://doi.org/10.1186/s12938-024-01212-4
<a id="4">[4]</a> Ahmed, S., Hasan, M. B., Ahmed, T., Sony, R. K., & Kabir, M. H. (2022). Less is More: Lighter and Faster Deep Neural Architecture for Tomato Leaf Disease Classification. IEEE Access, 10, 68868–68884. https://doi.org/10.1109/access.2022.3187203
<a id="5">[5]</a> Thakur, P. S., Khanna, P., Sheorey, T., & Ojha, A. (2022). Vision Transformer for plant Disease Detection: PlaNtVIT. In Communications in computer and information science (pp. 501–511). https://doi.org/10.1007/978-3-031-11346-8_43
<a id="6">[6]</a> Jain, A., & Biswas, S. (2023). Deep learning based music recommendation system using big data analytics. Journal of Big Data, 10(1), 1-17. https://doi.org/10.1186/s40537-023-00863-9
<a id="7">[7]</a> He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770–778). https://ieeexplore.ieee.org/document/7780459
<a id="8">[8]</a> Google Research. (n.d.). ViT-Base-Patch16-224-in21k. Hugging Face. Retrieved June 11, 2024, from https://huggingface.co/google/vit-base-patch16-224-in21k
<a id="9">[9]</a> WellCh4n. (n.d.). Tomato Leaf Disease Image Dataset. Hugging Face. Retrieved June 11, 2024, from https://huggingface.co/datasets/wellCh4n/tomato-leaf-disease-image
<a id="10">[10]</a> Maurício, J., Domingues, I., & Bernardino, J. (2023). Comparing vision Transformers and convolutional neural Networks for image classification: A literature review. Applied Sciences, 13(9), 5521. https://doi.org/10.3390/app13095521
<a id="11">[11]</a> Microsoft. (n.d.). ResNet-50. Hugging Face. Retrieved June 11, 2024, from https://huggingface.co/microsoft/resnet-50
<a id="12">[12]</a> Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, & Neil Houlsby. (2021). An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. https://arxiv.org/abs/2010.11929
<a id="13">[13]</a> Cheng, K.-T., & Lin, Y.-C. (2024). Development of skip connections in deep neural networks for computer vision and medical image analysis: A survey. arXiv. https://ar5iv.labs.arxiv.org/html/2405.01725v1
<a id=14>[14]</a> Bichen Wu, Chenfeng Xu, Xiaoliang Dai, Alvin Wan, Peizhao Zhang, Zhicheng Yan, Masayoshi Tomizuka, Joseph Gonzalez, Kurt Keutzer, & Peter Vajda. (2020). Visual Transformers: Token-based Image Representation and Processing for Computer Vision. https://arxiv.org/abs/2006.03677
<a id=15>[15]</a> Grigolli, J. F. J., Kubota, M. M., Alves, D. P., Rodrigues, G. B., Cardoso, C. R., Da Silva, D. J. H., & Mizubuti, E. S. G. (2011). Characterization of tomato accessions for resistance to early blight. Crop Breeding and Applied Biotechnology, 11(2), 174–180. https://doi.org/10.1590/s1984-70332011000200010