# BIDS Derivatives
## Background
### BIDS
Neuroimaging experiments result in complicated data that can be arranged in many different ways. The [Brain Imaging Data Structure] (BIDS, [[2]]) is a comprehensive and use-case-driven way of organizing neuroimaging and behavioral data.
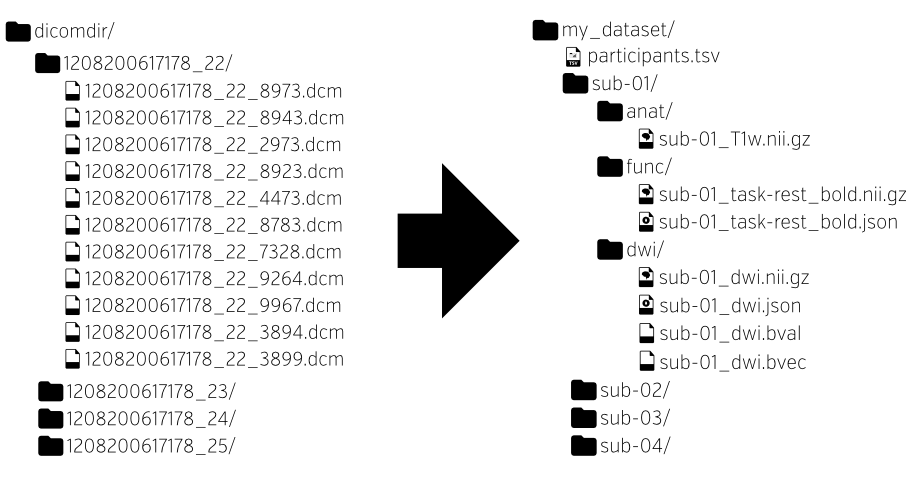
Originally written for MRI studies, BIDS has added descriptions for organizing electrophysiological (EEG [[8]], MEG [[7]] and iEEG/ECoG [[6]]) data. Work is being done to add PET, ASL and NIRS to the standard, among other modalities.
### BIDS Apps
A common specification of neuroimaging datasets affords queries for and adaptation to the available data. [BIDS Apps] are programs that accept BIDS data as inputs, and produce some output. This permits a simple command-line protocol:
```
bids-app /bids-directory /output-directory participant [OPTIONS]
```
There are a growing number of data analysis software packages that can understand data organised according to BIDS.

The output of a BIDS App is a derivative of the input dataset. BIDS Derivatives seeks to formalize this notion.
## BIDS Derivatives
Derivatives are outputs of (pre-)processing pipelines, capturing data and meta-data sufficient for a researcher to understand and (critically) reuse those outputs in subsequent processing. Standardizing derivatives is motivated by use cases where formalized machine-readable access to processed data enables higher level processing.
A derivative dataset is a collection of derivatives, or files that have been generated from the data. Broadly, a derivative can be considered to be preprocessed or processed, such that the data type is unchanged or changed, respectively, from that of the source data file(s).
BIDS Derivatives was finalized in version 1.4.0 of the BIDS specification.
### Tour of a BIDS Derivative
As with BIDS datasets, all conformant derivative datasets contain a `dataset_description.json`. New fields include `DatasetType`, which distinguishes `"derivative"` datasets from `"raw"`; `GeneratedBy`, a list of processes that generated the data; `SourceDatasets`, a list of datasets used to generate the derivative.
```YAML
{
"Name": "FMRIPREP Outputs",
"BIDSVersion": "1.4.0",
"DatasetType": "derivative",
"GeneratedBy": [
{
"Name": "fmriprep",
"Version": "1.4.1",
"Container": {
"Type": "docker",
"Tag": "poldracklab/fmriprep:1.4.1"
}
},
{
"Name": "Manual",
"Description": "Re-added RepetitionTime metadata to bold.json files"
}
],
"SourceDatasets": [
{
"DOI": "10.18112/openneuro.ds000114.v1.0.1",
"URL": "https://openneuro.org/datasets/ds000114/versions/1.0.1",
"Version": "1.0.1"
}
]
}
```
#### Preprocessed data
Data is considered to be *preprocessed* if it is fundamentally similar to the source data. Artifact removal, motion correction and resampling to a template space are examples of preprocessing.
An example of a subject with simultaneous EEG/fMRI resting state scan, aligned along with a T1w image to the MNI305 template:
```
pipeline1/
sub-01/
anat/
sub-01_space-MNI305_T1w.nii.gz
sub-01_space-MNI305_T1w.json
eeg/
sub-01_task-rest_desc-filtered_eeg.edf
sub-01_task-rest_desc-filtered_eeg.json
func/
sub-01_task-rest_space-MNI305_desc-preproc_bold.nii.gz
sub-01_task-rest_space-MNI305_desc-preproc_bold.json
```
The `space` entity indicates that a file is aligned to some reference space. For standard templates, this is sufficient. For custom templates (e.g., individual or study-specific), additional `SpatialReference` metadata is required in the JSON sidecar files.
The `desc` (description) entity allows for unrestricted alphanumeric labels, in the absence of a more appropriate entity to distinguish one file from another.
#### Derivative data types
Data is considered to be *processed* if it is fundamentally different to the source data. Processed data may differ substantially in BIDS datatypes from the original input data.
The initial offering of BIDS Derivatives only specifies anatomical derivatives that are of general use: masks and segmentations.
Mask images are binary images with 1 representing the region of interest and all other voxels containing 0. The following example shows a manually constructed lesion mask:
```
manual_masks/
sub-01/
anat/
sub-01_desc-lesion_mask.nii.gz
sub-01_desc-lesion_mask.json
```
A mask of the functionally-defined area fusiform face area could be encoded:
```
localizer/
sub-01/
func/
sub-01_task-loc_space-individual_label-FFA_mask.nii.gz
sub-01_task-loc_space-individual_label-FFA_mask.json
```
BIDS Derivatives introduces "discrete segmentations" and "probabilisitic segmentations".
> A *segmentation* is a labeling of regions of an image such that each location (for example, a voxel or a surface vertex) is identified with a label or a combination of labels. Labeled regions may include anatomical structures (such as tissue class, Brodmann area or white matter tract), discontiguous, functionally-defined networks, tumors or lesions.
>
> A *discrete segmentation* represents each region with a unique integer label. A *probabilistic segmentation* represents each region as values between 0 and 1 (inclusive) at each location in the image, and one volume/frame per structure may be concatenated in a single file.
A BIDS App that calculates ROIs in BOLD space from the automated anatomical labeling (AAL, doi:[10.1006/nimg.2001.0978]) could store discrete and probabilistic (or partial volume) segmentations as follows:
```
tissue_segmentation/
desc-AAL_dseg.tsv
desc-AAL_probseg.json
sub-01/
func/
sub-01_task-rest_desc-AAL_dseg.nii.gz
sub-01_task-rest_desc-AAL_probseg.nii.gz
```
The `dseg.tsv` file is a lookup table for interpreting a discrete segmentation and `probseg.json` contains a list identifying the labels for each consecutive volume.
#### Unspecified data types
Derivatives can never be fully specified, as new methods can always be developed, requiring new data representations. BIDS recognizes this and encourages adopting "BIDS-style naming conventions":
> Additional files and folders containing raw data MAY be added as needed for special cases. All non-standard file entities SHOULD conform to BIDS-style naming conventions, including alphabetic entities and suffixes and alphanumeric labels/indices. Non-standard suffixes SHOULD reflect the nature of the data, and existing entities SHOULD be used when appropriate.
This recommendation remains in force for derivatives datasets. Additionally, BIDS Derivatives acknowledges that it may be desirable to distribute derivatives generated by non-compliant applications, for the sake of reproducibility and non-duplication of effort. Therefore,
> if a BIDS dataset contains a `derivatives/` sub-directory, the contents of that directory may be a heterogeneous mix of BIDS Derivatives datasets and non-compliant derivatives.
One example of such a non-compliant derivative dataset would be FreeSurfer reconstructions of subject surfaces:
```
bids-root/
derivatives/
freesurfer/
sub-01/
label/
mri/
...
...
sub-01/
anat/
sub-01_T1w.nii.gz
...
```
Note that subject directory names conform to BIDS conventions, but contents are determined by the generating application, in this case, FreeSurfer.
### Organizing datasets and their derivatives
BIDS Derivatives datasets are intended to be interpretable and distributable with or without the datasets used to generate them. This is necessary for storage and bandwidth constraints, as well as to permit the distribution of derivatives when the source data are restricted.
This independence affords flexibility in the relative organization of datasets. The following examples show three ways to organize, relative to each other, a raw BIDS dataset, a preprocessed derivative dataset, and an analysis that uses both as inputs.
A collection of derivative datasets may be stored in the `derivatives/` subdirectory of a BIDS (or BIDS Derivatives) dataset:
```
my_dataset/
derivatives/
preprocessed/
analysis/
sub-01/
...
```
A BIDS Derivatives dataset may contain references to its input datasets in the `sourcedata/` subdirectory:
```
my_analysis/
sourcedata/
raw/
preprocessed/
sub-01/
...
```
Note that the `sourcedata/` and `derivatives/` subdirectories constitute dataset boundaries. Any contents of these directories may be validated independently, but their contents must not affect the interpretation of the nested or containing datasets.
Unnested datasets are also possible. For example:
```
my_study/
raw_data/
sub-01/
...
derivatives/
preprocessed/
analysis/
```
## Learn More
The full BIDS specification is available at **[bids-specification.readthedocs.io](https://bids-specification.readthedocs.io)**. Self-contained PDFs are archived on Zenodo (**doi:[10.5281/zenodo.3686061]**).
The **[BIDS Starter Kit](https://github.com/bids-standard/bids-starter-kit)** is a more informal, human-friendly introduction to BIDS.
## Get Involved
BIDS is a collaborative effort, and contributions of all kinds are welcome!
The NeuroStars forum (https://neurostars.org) is a forum to ask, search for and answer questions about any neuroscience topic, and the BIDS community strongly recommends this resource. For BIDS-specific questions, the `bids` tag (https://neurostars.org/tag/bids) makes your question easier to find.
The BIDS specification can be extended in a backwards compatible way and will evolve over time. These are accomplished with BIDS Extension Proposals (BEPs), which are community-driven processes. A list of BEPs, as well as instructions on how to propose a new BEP, can be found at https://bids.neuroimaging.io/get_involved.html.
If the specification is ambiguous, inconsistent or silent on some point, proposals can be made to the BIDS Specification (https://github.com/bids-standard/bids-specification/) GitHub repository. The BIDS Starter Kit (https://github.com/bids-standard/bids-starter-kit/) repository exists to provide a more user-friendly guide, and accepts proposals for improvement, as well.
## The Making Of BIDS Derivatives
The need to specify BIDS Derivatives was identified during the early stages of BIDS specification, and a BIDS Extension Proposal (BEP) was started in February 2016, prior to the release of BIDS 1.0 in June 2016.
Development of the proposal was largely based on the experience of developing BIDS Apps. As multiple applications produced similar or equivalent derivatives, common naming schemes were added to the proposal to facilitate reuse of the derivatives. For example, the Configurable Pipeline for the Analysis of Connectomes (C-PAC) and fMRIPrep took similar inputs and had a broad overlap in their outputs, and so made sense to coordinate.
An August 2017 meeting at Stanford led to an agreement to divide the increasingly large BEP into a series of BEPs, most focused on particular modalities or use cases.
In July 2018, a survey of the neuroimaging community was taken to establish priorities (essential, desirable or inessential) for structural, functional and diffusion MRI derivatives. The results of the survey[[1]] were posted in advance of an August 2018 workshop of 31 participants, where sub-proposals were pushed toward completion and common principles were established. In December 2018, Release Candidate 1 was published, including all imaging modalities, for implementation and feedback.
In July 2019, a “Common Derivatives” proposal was re-introduced establishing more general principles, to be followed by subsequent modality-specific and non-imaging proposals. Common Derivatives entered final review in May 2020 and were released as part of BIDS 1.4.0 in June 2020.
## Future Directions
The initial offering of BIDS Derivatives is intended to establish a set of ground rules for future elaboration.
There are existing BIDS Extension Proposals (BEPs) for the following derivatives:
* Structural MRI derivatives ([BEP011])
* Functional MRI derivatives ([BEP012])
* Diffusion MRI derivatives ([BEP016])
* Affine transformations and nonlinear warp fields ([BEP014])
* Connectivity data schema ([BEP017])
* Common electrophysiological (EEG/MEG/iEEG) derivatives ([BEP021])
* PET preprocessing derivatives ([BEP023]])
* Provenance ([BEP028])
Statistical and computational modeling derivatives are a logical further effort, and are likely to result in BEPs in the near future.
## References
[[1]] Feingold, F.W. (2018), 'BIDS-Processed Data Survey Results', Stanford Center for Reproducible Neuroscience, http://reproducibility.stanford.edu/bids-processed-data-survey-results/
[[2]] Gorgolewski, K.J. (2016), 'The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments' Scientific Data, 3:160044. doi:10.1038/sdata.2016.44
[[3]] Gorgolewski, K.J. (2017a), 'BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods', PLOS Computational Biology 13(3): e1005209, doi:10.1371/journal.pcbi.1005209
[[4]] Gorgolewski K.J. (2017b), 'OpenNeuro – a free online platform for sharing and analysis of neuroimaging data [version 1; not peer reviewed]’, F1000Research, 6:1055 (poster), doi:10.7490/f1000research.1114354.1
[[5]] Gorgolewski, K.J. (2017c), 'Restructuring BIDS Derivatives', bids-discussion mailing-list, https://groups.google.com/forum/#!msg/bids-discussion/l74eKXzNX84/4rMQzS_KAwAJ
[[6]] Holdgraf, C. (2019), 'iEEG-BIDS, extending the Brain Imaging Data Structure specification to human intracranial electrophysiology' Scientific Data, 6:102. doi:10.1038/s41597-019-0105-7
[[7]] Niso, G. (2018), 'MEG-BIDS, the brain imaging data structure extended to magnetoencephalography' Scientific Data, 5:180110. doi:doi:10.1038/sdata.2018.110
[[8]] Pernet, C.R. (2019), 'EEG-BIDS, an extension to the brain imaging data structure for electroencephalography' Scientific Data, 6:103. doi:10.1038/s41597-019-0104-8
[[9]]: https://bids-specification.readthedocs.io/en/stable/
[[10]]: https://nda.nih.gov/
[1]: http://reproducibility.stanford.edu/bids-processed-data-survey-results/
[2]: https://www.nature.com/articles/sdata201644
[3]: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005209
[4]: https://f1000research.com/posters/6-1055
[5]: https://groups.google.com/d/msg/bids-discussion/l74eKXzNX84/4rMQzS_KAwAJ
[6]: https://www.nature.com/articles/s41597-019-0105-7
[7]: https://www.nature.com/articles/sdata2018110
[8]: https://www.nature.com/articles/s41597-019-0104-8
[9]: https://bids-specification.readthedocs.io/en/stable/
[10]: https://nda.nih.gov/
[10.5281/zenodo.3686061]: https://doi.org/10.5281/zenodo.3686061
[10.1006/nimg.2001.0978]: https://doi.org/10.1006/nimg.2001.0978
[Brain Imaging Data Structure]: https://bids-specification.rtfd.io/
[BIDS Apps]: https://bids-apps.neuroimaging.io/apps/
[BIDS Apps paper]: https://doi.org/10.1371/journal.pcbi.1005209
[BEP011]: https://bids.neuroimaging.io/bep011
[BEP012]: https://bids.neuroimaging.io/bep012
[BEP013]: https://bids.neuroimaging.io/bep013
[BEP014]: https://bids.neuroimaging.io/bep014
[BEP015]: https://bids.neuroimaging.io/bep015
[BEP016]: https://bids.neuroimaging.io/bep016
[BEP017]: https://bids.neuroimaging.io/bep017
[BEP018]: https://bids.neuroimaging.io/bep018
[BEP019]: https://bids.neuroimaging.io/bep019
[BEP020]: https://bids.neuroimaging.io/bep020
[BEP021]: https://bids.neuroimaging.io/bep021
[BEP022]: https://bids.neuroimaging.io/bep022
[BEP023]: https://bids.neuroimaging.io/bep023
[BEP024]: https://bids.neuroimaging.io/bep024
[BEP025]: https://bids.neuroimaging.io/bep025
[BEP026]: https://bids.neuroimaging.io/bep026
[BEP027]: https://bids.neuroimaging.io/bep027
[BEP028]: https://bids.neuroimaging.io/bep028
[BEP029]: https://bids.neuroimaging.io/bep029
[BEP030]: https://bids.neuroimaging.io/bep030