# Hüden Turgut - Influenza Viruses
# ****Influenza Viruses****
Influenza is generally known as “the flu,” is infected by an influenza virus. Influenza are found within the Orthomyxoviridae family.
---
# Influenza Types

**Table 10.1 Types of Influenza Viruses**
Three genera exist with this family that pertain to influenza viruses, Influenzavirus A, Influenzavirus B, and Influenzavirus C, each of which contain a single species, or type: Influenza A virus, Influenza B virus, and Influenza C virus, respectively (Table 10.1).
(*source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.171)
---
# Host Range of Influenza Viruses
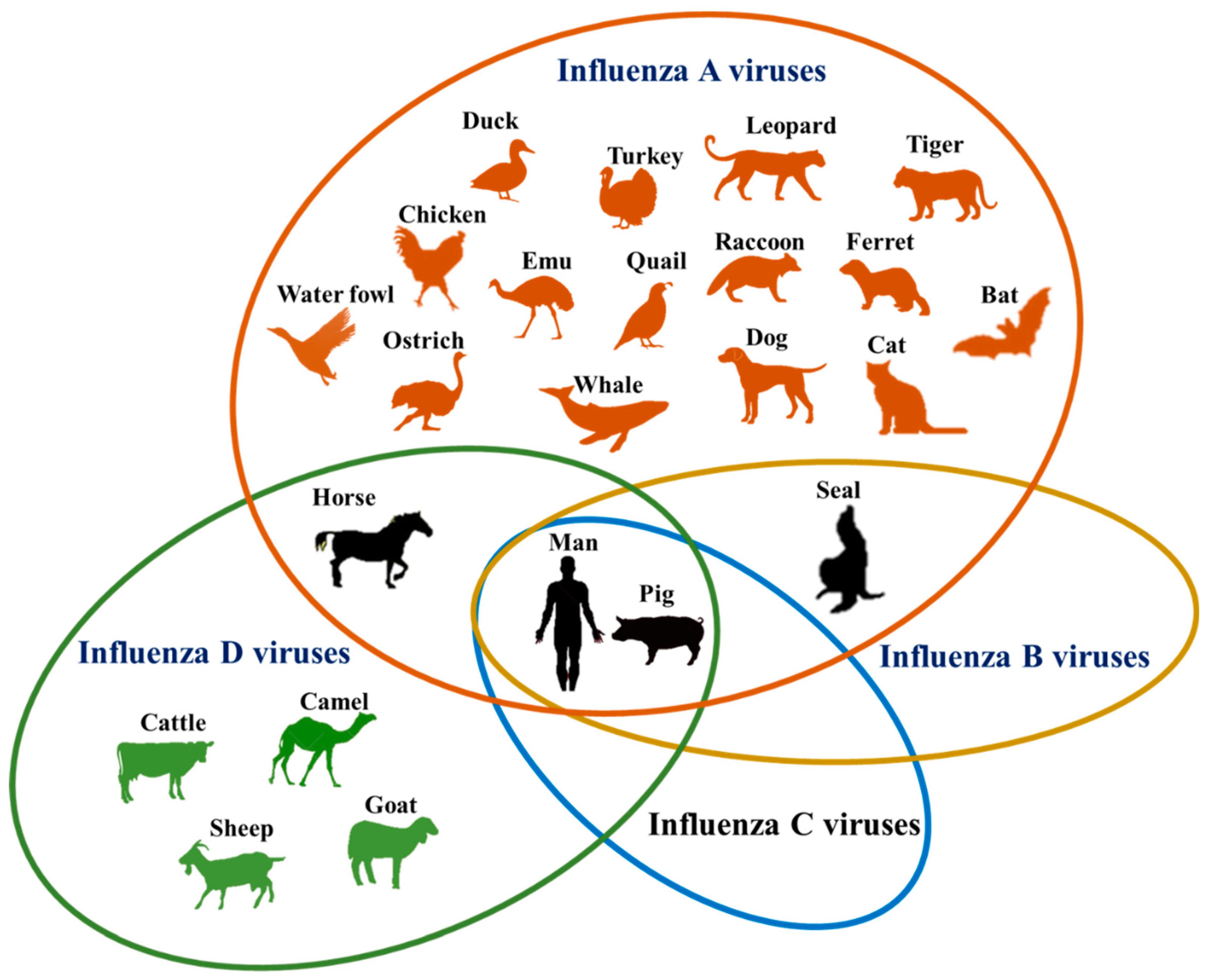
Source: [wikipedia](https://en.wikipedia.org/wiki/Orthomyxoviridae)
-----
# Influenza Infected Area

***Figure 10.1 Influenza-susceptible areas of the respiratory tract.***
----
>* Influenza is transmitted to humans through the respiratory route.
>* Human influenza viruses infect the ciliated columnar epithelium found in the sinuses, nasopharynx, larynx, trachea, bronchi, and bronchioles.
(*source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.173)
----
# Influenza Virus Virion

----
>* Influenza virions are enveloped and contain a helical −ssRNA genome, referred to as the viral RNA (vRNA).
>* Virions are spherical, and about 80–120 nm in diameter.
>*The viral RNAs that make up the genome are shown as red coils and bound to ribonucleoproteins (RNP).
Source : [wikipedia](https://en.wikipedia.org/wiki/Influenza)
-----
# Influenza A Virion Structure

***Figure 10.3 The structure of influenza A virions.***
---
>* The influenza genome is segmented. Each helical segment encodes a different viral gene.
>* Influenza A and B have eight gene segments(that encode 11 proteins), while influenza C has seven (that encode 9 proteins).
>* Two integrated glycoproteins, HA (Hemagglutinin) and NA (Neuraminidase), embedded in the influenza envelope.
>* HA protein is 4 times more than NA.
(*source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.175)
---
# Influenza Gene Size and Functions

***Figure 10.4 Size and function of influenza genes.***
---
>* The influenza A genome includes eight segments that encode 11 proteins.
>* The gene segments are ordered from largest to smallest.
>* Small protein PB1-F2 is derived from an alternative reading frame in some strains.
>* M2 and NS2/NEP are products of alternative splicing.
(*source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.176)
---
# Influenza Viruses Receptor

***Figure 10.5A Sialic acid, the receptor for influenza viruses.***
---
>* Influenza is a respiratory virus that attaches to the ciliated respiratory epithelium within the nasopharynx and trachea, bronchi, and bronchioles (the tracheobronchial tree).
>* The HA1 subunit of the HA protein binds to carbohydrates extending from the plasma membrane of ciliary lung cells.
>* The influenza HA protein is attached to a sugar known as sialic acid, which is located at the very end of these carbohydrate chains.
(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.176)
---
# Attachment, Penetration and Fusion

***Figure 10.6A Influenza virus attachment, penetration, and uncoating.***
----
>* The influenza HA protein attaches to terminal sialic acids of membrane glycoproteins, and the virion enters the cells via clathrin-mediated endocytosis.
>* The HA protein changes configuration within the acidified endosome, leading to the fusion of the viral envelope with the endosomal membrane.
>* The M2 protein acts as a channel to allow H+ ions to enter the virion, further weakening the virion structure.
>* The eight RNPs are released into the cytoplasm.
(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.178)
-----

***Figure 10.6B Influenza virus attachment, penetration, and uncoating.***
----
>**1.** The HA protein is composed of two subunits, HA1 and HA2 that form a trimer.
>**2.** HA1 facilitates attachment to sialic acid, but in the acidified endosome, the HA1 undergoes a conformational change.
>**3.** The HA2 subunits are translocated toward the endosomal membrane, where the HA2 fusion peptides (asterisk) make contact.
>(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.178)
----

***Figure 10.6C Influenza virus attachment, penetration, and uncoating.***
----
>**4.** After the endosome is acidified and the HA2 fusion peptides engage the endosomal membrane, the molecule folds back upon itself, drawing the two membranes together.
>**5.** A channel is created through which RNPs are exported.
>(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.178)
------
# Replication

***Figure 10.7 Initiation of transcription via cap-snatching.***
------
>* The influenza RdRp complex steals the 5′ caps of host mRNA transcripts for its own vmRNA transcripts.
>* PB2 binds the host mRNA 5′ cap, which PA cleaves off, leaving the host mRNA to become degraded. PB1 extends the nucleotides of the capped sequence, using the vRNA as a template.
>* When it reaches the end of the vRNA segment, it encounters a series of uridines on which it stutters, creating the poly(A) tail of the vmRNA transcript.
>* This transcription process is repeated for all eight segments of the genome.
>(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.178)
------
# Assembly, Maturation, and Release

***Figure 10.8 Assembly, maturation, and release.***
-------
>* Following translation, HA, NA, and M2 localize to the plasma membrane. M1 recruits the RNPs to the budding virion.
>* As the enveloped virion is released, NA cleaves sialic acids from plasma membrane proteins or from the HA protein to prevent aggregation of virions at the cell surface.
>(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.179)
------
# Influenza Pandemic
Influenza virus caused pandemic in 1918 and 2009 years.The H3N2 and H1N1 viruses are the current seasonal human influenza A subtypes.

***Figure 10.12 Pandemics of the 20th century caused by antigenic shift.***
**Spanish flu pandemic** The 1918 was a particularly virulent strain that spread in waves throughout the world, leading to the deaths of an estimated 20–50 million people worldwide.
**Asian flu** In 1957, antigenic shift occurred as the H1N1 virus was replaced with the H2N2 subtype, a reassortant with avian influenza viruses.
**Hong Kong flu** Another antigenic shift, to H3N2, occurred in 1968.
**1977 Russian flu** The H1N1 influenza reappeared in 1977 and began circulating alongside H3N2 in the human population.
**Swine flu pandemic** In 2009, a quadruple-reassortant H1N1 virus containing vRNA segments from avian, human, and swine influenza viruses began infecting the human population. The 2009 H1N1 “swine flu” was not as virulent as the 1918 H1N1 strain, and a third of people over age 60 possessed neutralizing antibodies against it, suggesting a similar strain had circulated previously in the human population.
>(*Source*: Louten, J. (2016). Influenza Viruses. Essential Human Virology,p.183)
-------
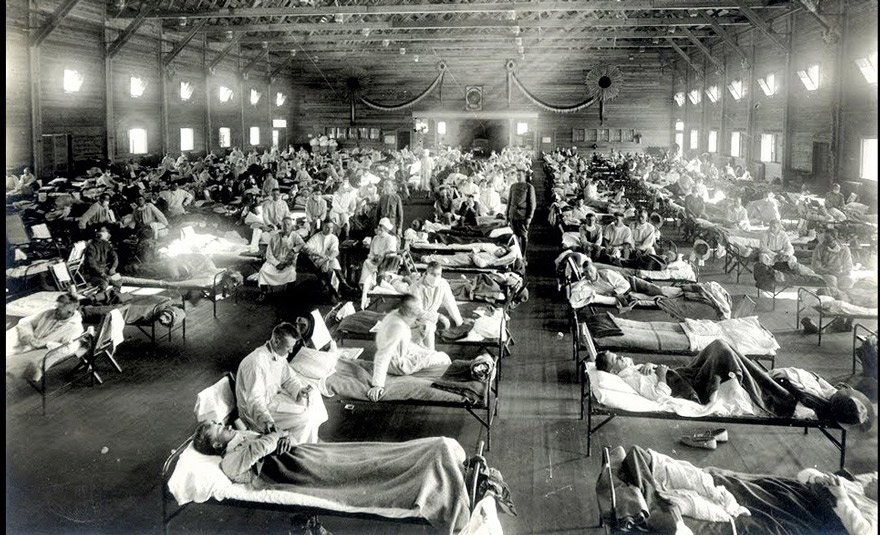
Photo from the 1918 pandemic. The same situation is unfortunately experienced by covid 19.
Source: [sözcü](https://www.sozcu.com.tr/2020/yazarlar/sinan-meydan/savaslardan-cok-salginlar-oldurdu-ispanyol-gribi-5681024/)