# Reoviruses (*Reoviridae*)
---
## 1.) Reoviridae
---
| **Feature** | **Reoviridae** |
|:------------------:|:------------------------:|
| Genetic Material | dsRNA |
| Type of Genetic Material | Segmented, all segments are linear in themselves |
| Type of Symmetry | Icosahedral |
| Envelope Existence | Non-enveloped |
| Host Diversity | All living things except bacterias (archaebacteria & eubacteria) |
| Unique Feature | Multi capsid layers |
| Most Famous One | Rotavirus |
| **Rotavirus Disease** | **Diarrhea** |
:::info
<p style="text-align: center"> Rotavirus </p>
:::
<img src="https://viralzone.expasy.org/resources/Rotavirus_virion.jpg" alt="Reoviridae" title="Reoviridae Struture" width="750" height="" />
---

---
## 2.) Basics of Reoviruses
- The family Reoviridae is large and diverse, containing **two subfamilies** and numerous genera (*Spinareovirinae and Sedoreovirinae*).
- The best-known reovirus is **Rotavirus**, **Orbivirus**, **Orthoreovirus**, and **Fijivirus**.
- The family name Reoviridae comes from **respiratory** and **enteric** viral diseases. However, these diseases were caused by **orphanviruses**, not Reoviruses, which were thought to be.
:::info
**An orphanvirus is a virus that is not associated with a disease but may possess pathogenicity.**
:::
- Virions are **unique having two or three discrete capsid layers**. The innermost capsid layer **encloses dsRNA segments and enzymes** for RNA synthesis because of host's protective actions.
:::info
**In any living species, dsRNA cannot be considered normal. This is an abnormality or a disease according to them. Defense systems defined to this break down the dsRNA. This layered capsid system is the Reoviruses' self-protection system.**
:::
---
## 3.) Genome Organization
<img src="https://viralzone.expasy.org/resources/Reoviridae_genome.jpg" alt="Rotavirus Genome" title="Rotavirus Genome" width="550" height="" />
:::success
Reovirus genomes contain from 9 to 12 segments of linear, dsRNA. For the most part, each segment encodes a single protein. For example, Rotavirus and Orbivirus genomes have 11 segments encoding 12 proteins. Genome segment sizes range from 600 to 3000 bp for a total coding capacity of approximately 18,500-19,200 bp.
:::
---
## 4.) Virus Attachment
<img style="float: right;" src="https://i.imgur.com/R9lfzuX.jpg" alt="Rotavirus Attachment" title="Rotavirus Attachment" width="350" height="" />
:::success
- Reovirus attachment is mediated by capsid proteins (Rotavirus VP4, Orthoreovirus σ1, Orbivirus VP2). A variety of cell surface molecules act as receptors.
- Some Reoviruses use protein receptors, others use carbohydrate receptors (for example, sialic acid) and some use a combination of the two. Rotavirus VP4 is glycosylated and is cleaved by extracellular proteases found in the digestive tract.
- Rotavirus VP4 binds to histo blood group antigens (HGBAs) complex carbohydrates found on epithelia of the respiratory, genitourinary, and digestive tracts.
:::
---
## 5.) Virus Entry
<img style="float: right;" src="https://i.imgur.com/zePnhWm.jpg" alt="Rotavirus Entry" title="Rotavirus Entry" width="" height="" />
:::success
- Reoviruses encode proteins that interact with membranes to form pores or channels (Rotavirus NSP1, Orbivirus VP5, Orthoreovirus μ1 protein).
- Penetration across plasma membrane or endosomal membranes is accompanied by disassembly of the outer capsid layers.
- The innermost core, containing genome segments, undergoes molecular rearrangements, but remains intact.
- In the case of Rotaviruses, disassembly of outer layers is triggered by the low calcium concentration of the cytosol.
:::
---
## 6.) Virus Transcription
<img style="float: right;" src="https://i.imgur.com/p1txjG9.jpg" alt="Rotavirus Transcription" title="Rotavirus Transcription" width="" height="" />
:::success
- Penetration and uncoation activate the transcription activities of the core. Viral mRNA is synthesized within the core and newly formed (capped) transcripts are released through pores found at the vertices of the inner capsid.
- It is assumed that each genome segment is associated with its own pore and transcription complex. mRNAs are not polyadenylated, but form a hairpin structure at the 3' end.
- In the case of Rotaviruses, synthesis of viral transcripts is mediated by a complex of VP1 and VP3. VP1 contains the catalytic RNA synthesis domain while VP3 binds single stranded RNA (ssRNA) and has nucleotide phosphohydrolase, guanyltranserase, and methyltransferase activities (thus VP3 is the cap-synthesizing component of the transcription complex).
:::
:::info
**Transcriptionally active cores can be prepared from purified virions (by removal of the outer capsid layers) and in vitro these can produce up to milligram quantities of mRNA, given an adequate source of precursors.**
:::
---
## 7.) Virus Translation
<img style="float: right;" src="https://i.imgur.com/ljyyh0L.jpg" alt="Rotavirus Translation" title="Rotavirus Translation" width="" height="" />
:::success
- Upon exiting the cores, mRNAs are translated. In the case of Rotaviruses, nonstructural protein 3 (NSP3) binds to "eIF4F", the cytoplasmic cap binding complex to facilitate translation. Thus NSP3 seems to serve the function of host polyA-binding protein in translation initiation.
- Some Rotavirus mRNAs are translated in the cytosol but mRNAs for rotavirus VP4, VP7, and NSP4 are translated on rough endoplasmic reticulum (ER), and the proteins are glycosylated therein.
- Because of their segmented genomes, Reoviruses do not carry a separate gene sequence for each protein. Instead of carrying a separate gene for each protein, they carry some proteins in any other available gene segment. So how do they get these parts read into the ribosome? Here, the system they made is called "leaky scanning".
:::
---
### -) Leaky Scanning
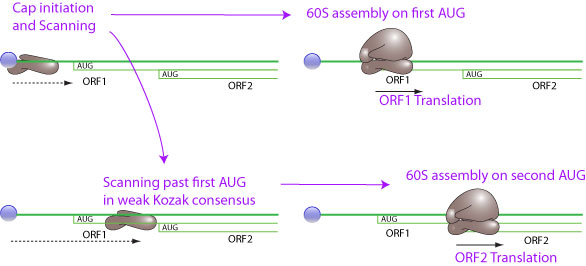
:::info
**Leaky scanning is a phenomenon in which a weak initiation codon triplet on mRNA is sometimes skipped by ribosome in translation initiation. The 40S ribosomal subunit continues scanning to further initiation codon. The weak initiation codon can be an ACG, or an ATG in a weak Kozak consensus context. This way an mRNA can encode for several different proteins if the AUG are not in frame, or for proteins with different N-terminus if the AUG are in the same frame.**
:::
---
## 8) Virus Replication
<img style="float: right;" src="https://i.imgur.com/LHbLN7n.jpg" alt="Rotavirus Genome Replc. & Assembly" title="Rotavirus Genome Replc. & Assembly" width="" height="" />
:::success
- Cytosolic proteins form dense viral organelle-like structures called virus factories or the viroplasm. Cores assemble in the viroplasm and these are capable of transcription to generate additional mRNAs.
- It is clear that each mRNA is the template for synthesis of single complementary (negative) strand to form a double stranded genome segment. This implies synthesis of complementary (-) RNA and initial steps of viral morphogenesis. Late transcription (genome replication) occurs in these progeny cores.
- Viroplasm are electronic dense cytoplasmic inclusions where viral replication and assembly take place. They are produced by Nucleo-Cytoplasmic Large DNA viruses like Poxviridae, Asfarviridae and Iridoviridae, dsRNA viruses like Reoviridae and ssRNA(-) viruses like Filoviridae.
:::
---
## 9) Virus Assembly
<img style="float: right;" src="https://i.imgur.com/LHbLN7n.jpg" alt="Rotavirus Genome Replc. & Assembly" title="Rotavirus Genome Replc. & Assembly" width="" height="" />
:::success
- Rotavirus double layer particles (DLPs) are synthesized in the viroplasm concurrent with genome replication. It is that pentamers of VP2 dimers (the inner core protein) interact with VP1 and VP3 and a single genome segment.
- Assembly of pentamers forms the inner core. Subsequently VP6 is added to form the DLP. Rotavirus DLPs then leave the viroplasm and bud into the ER lumen by virtue of interactions with molecules of NSP4 located in the ER membrane.
- Here, the packaging of gene segments is very important. Because one gene that transcribes each protein (or group of proteins) must come in.
:::
---
### -) Reoviridae Genome Packaging
:::info
**Segmented double-stranded dsRNA viruses share remarkable similarities in their replication strategy and capsid structure. Reoviridae rely on specific intermolecular interactions between +RNAs that guide multisegmented genome assembly.**
:::

- The proposed model of genome assortment and packaging in rotaviruses:
:::success
**1.** Within viroplasms, rotaviruses + RNAs bind viral RdRP (VP1, shown in red), and RNA capping enzyme (VP3, in blue), forming +RNA/VP1/VP3 complexes.
**2.** Binding of the octameric RNA-binding NSP2 (teal), causes structural remodeling of the viral +RNAs, exposing otherwise sequestered complementary sequences (sequences shown in red, blue and green). The complementary sequences promote base pairing between the different types of rotavirus +RNAs, a process representing RNA assortment.
**3.** The assorted RNA complex containing NSP2, VP1 and VP3, is predicted to nucleate VP2 core assembly.
**4.** In this model, core assembly results in the displacement of +RNA bound NSP2. RdRPs within new formed cores direct dsRNA synthesis, using +RNAs as templates (not shown).
:::
---
## 10) Virus Exit

:::success
- In this process, these core are coated with VP6, forming immature DLPs that bud across the membrane of the endoplasmic reticulum, acquiring a transient lipid membrane which is modified with the ER resident viral glycoproteins NSP4 and VP7; these enveloped particles also contain VP4. As the particles move towards the interior of the ER, the transient lipid membrane and the nonstructural protein NSP4 are lost, while the virus surface proteins VP4 and VP7 rearrange to form the outermost virus protein layer, yielding mature infectious triple-layered particles.
- The high Ca^+2^ concentration in the ER is necessary for assembly of VP4 and VP7 into the outmost shell. Virions are released from the cell either by lysis or by exocytosis.
:::
---
## 11) All Process

---