<!---
title: 'UC9.3 Transcriptomics metadata'
tags:
- FAIR cookbook
- UC9
- metadata
--->
# UC9.3 Transcriptomics metadata
[TOC]
## Main Objectives:
The main purpose of this recipe is:
> To provide guidance on the minimum set of metadata and semantics required to describe any transcriptomics experiment, from standard case-control to dosage-response designs, and from microarrays to single cell RNA sequencing.
### Who is this recipe aimed at?
This document is aimed at anyone interested in the metadata, and specifically the ontology annotations, required to capture variables and experimental factors related to a transcriptomics experiment. General knowledge of transcriptomics experiment would be beneficial. No specific technical knowledge is required.
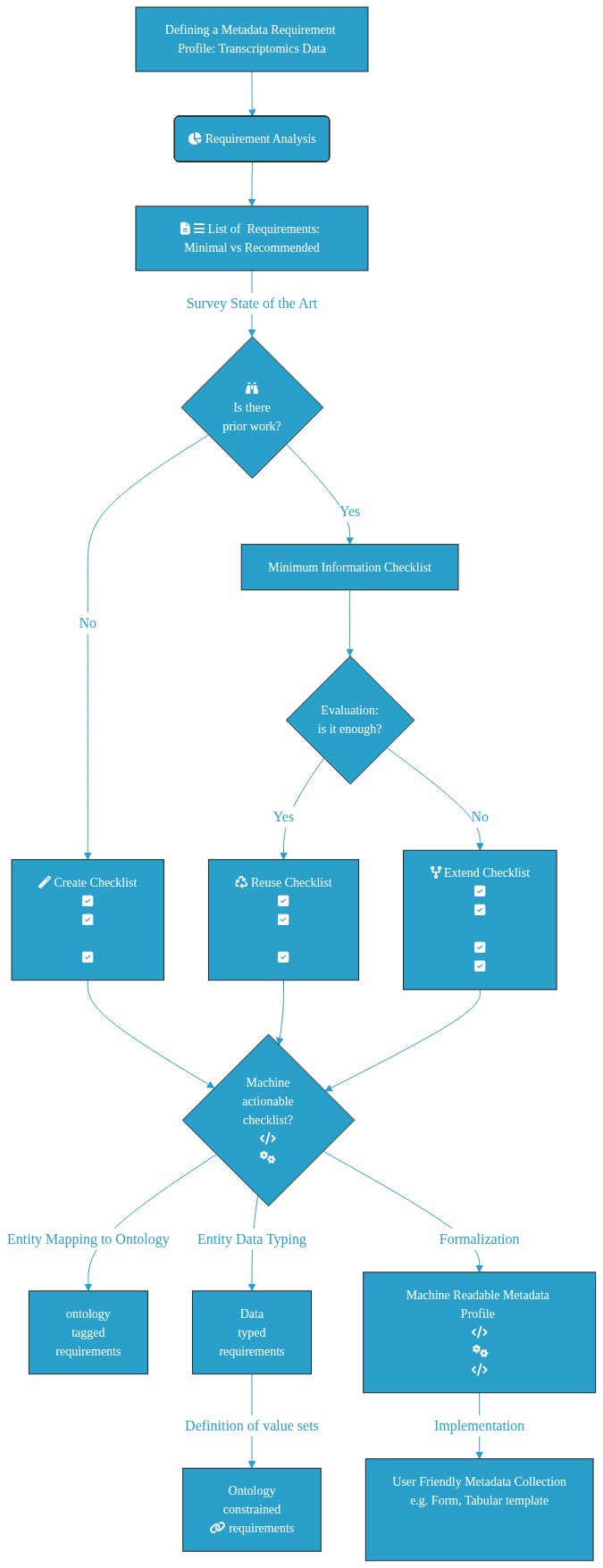
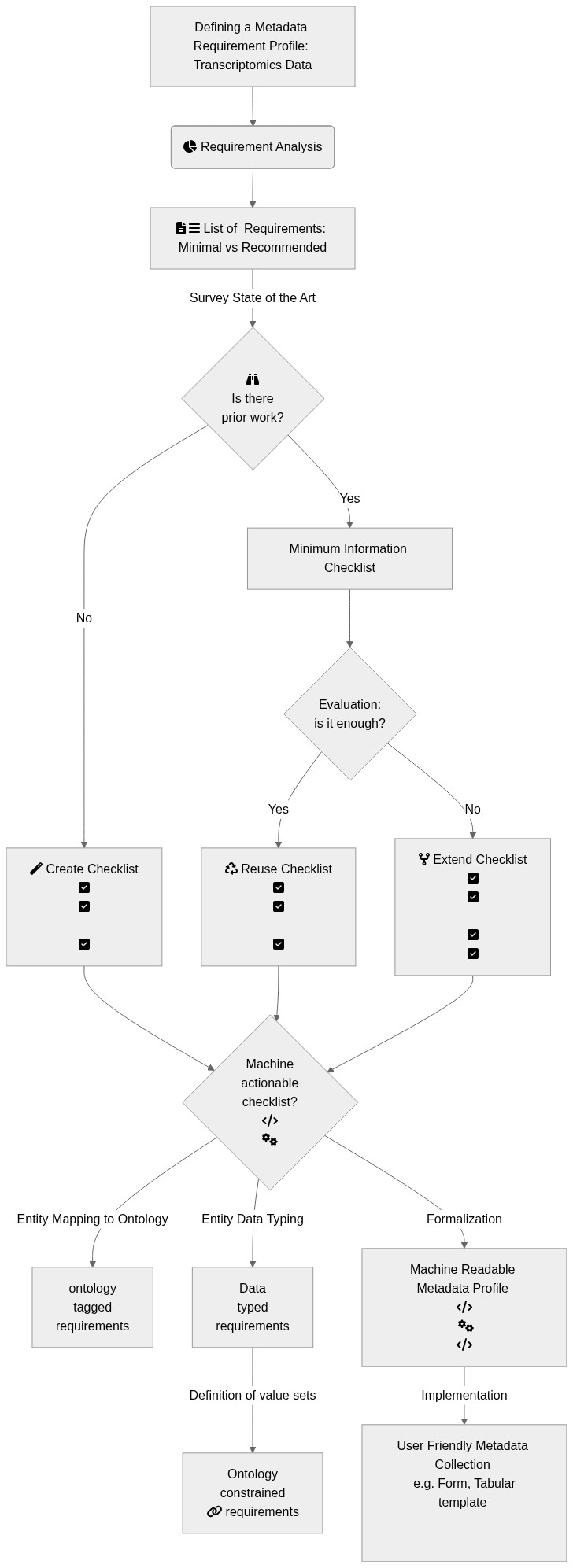
## Graphical Overview:
```
graph TD
A[Defining a Metadata Requirement Profile: Transcriptomics Data]:::box --> Z:::box
Z(fa:fa-pie-chart Requirement Analysis) --> W[fa:fa-file-text fa:fa-bars List of Requirements: Minimal vs Recommended]
W:::box --> |Survey State of the Art| C{fa:fa-binoculars <br> Is there<br> prior work?}:::box
C --> |No| E[fa:fa-magic Create Checklist <br>fa:fa-check-square<br>fa:fa-check-square<br>fa:fa-square-o<br>fa:fa-check-square]:::box
C --> |Yes| D[Minimum Information Checklist]:::box
D --> G{Evaluation:<br> is it enough?}:::box
G --> |Yes| H[fa:fa-recycle Reuse Checklist <br>fa:fa-check-square<br>fa:fa-check-square<br>fa:fa-square-o<br>fa:fa-check-square]:::box
G --> |No| I[fa:fa-code-fork Extend Checklist <br>fa:fa-check-square<br>fa:fa-check-square<br>fa:fa-square-o<br>fa:fa-check-square<br>fa:fa-check-square]:::box
H --> K{Machine<br> actionable<br> checklist?<br>fa:fa-code<br> fa:fa-cogs}:::box
E --> K{Machine<br> actionable<br> checklist?<br>fa:fa-code<br> fa:fa-cogs}:::box
I --> K{Machine<br> actionable<br> checklist?<br>fa:fa-code<br> fa:fa-cogs}:::box
K --> |Entity Mapping to Ontology| L[ontology<br> tagged <br>requirements]:::box
K --> |Entity Data Typing| M[Data<br> typed<br> requirements]:::box
M --> |Definition of value sets| N[Ontology<br> constrained<br> fa:fa-link requirements]:::box
K --> |Formalization| J[Machine Readable Metadata Profile <br>fa:fa-code<br> fa:fa-cogs<br>fa:fa-code]:::box
J --> |Implementation| O[User Friendly Metadata Collection <br> e.g. Form, Tabular template <br> fa:fa-file-excel-o fa:fa-file-excel-o fa:fa-file-excel-o<br> fa:fa-group fa:fa-group fa:fa-group]:::box
linkStyle 0,1,2,3,4,5,6,7,8,9,10,11,12,13,14,15 stroke:#2a9fc9,stroke-width:1px,color:#2a9fc9,font-family:avenir;
classDef box font-family:avenir,font-size:14px,fill:#2a9fc9,stroke:#222,color:#fff,stroke-width:1px
```
[](https://mermaid-js.github.io/mermaid-live-editor/#/edit/eyJjb2RlIjoiZ3JhcGggVERcbiAgQVtEZWZpbmluZyBhIE1ldGFkYXRhIFJlcXVpcmVtZW50IFByb2ZpbGU6IFRyYW5zY3JpcHRvbWljcyBEYXRhXTo6OmJveCAtLT4gWjo6OmJveFxuICBaKGZhOmZhLXBpZS1jaGFydCBSZXF1aXJlbWVudCAgQW5hbHlzaXMpIC0tPiBXW2ZhOmZhLWZpbGUtdGV4dCBmYTpmYS1iYXJzIExpc3Qgb2YgIFJlcXVpcmVtZW50czogTWluaW1hbCB2cyBSZWNvbW1lbmRlZF1cblxuICBXOjo6Ym94IC0tPiB8U3VydmV5IFN0YXRlIG9mIHRoZSBBcnR8IEN7ZmE6ZmEtYmlub2N1bGFycyA8YnI-IElzIHRoZXJlPGJyPiBwcmlvciB3b3JrP306Ojpib3hcblxuXG4gIEMgLS0-IHxOb3wgRVtmYTpmYS1tYWdpYyBDcmVhdGUgQ2hlY2tsaXN0IDxicj5mYTpmYS1jaGVjay1zcXVhcmU8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLXNxdWFyZS1vPGJyPmZhOmZhLWNoZWNrLXNxdWFyZV06Ojpib3hcbiAgQyAtLT4gfFllc3wgRFtNaW5pbXVtIEluZm9ybWF0aW9uIENoZWNrbGlzdF06Ojpib3hcbiAgRCAtLT4gR3tFdmFsdWF0aW9uOjxicj4gaXMgaXQgZW5vdWdoP306Ojpib3hcbiAgRyAtLT4gfFllc3wgSFtmYTpmYS1yZWN5Y2xlIFJldXNlIENoZWNrbGlzdCA8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLWNoZWNrLXNxdWFyZTxicj5mYTpmYS1zcXVhcmUtbzxicj5mYTpmYS1jaGVjay1zcXVhcmVdOjo6Ym94XG4gIEcgLS0-IHxOb3wgSVtmYTpmYS1jb2RlLWZvcmsgRXh0ZW5kIENoZWNrbGlzdCA8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLWNoZWNrLXNxdWFyZTxicj5mYTpmYS1zcXVhcmUtbzxicj5mYTpmYS1jaGVjay1zcXVhcmU8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlXTo6OmJveFxuICBIIC0tPiBLe01hY2hpbmU8YnI-IGFjdGlvbmFibGU8YnI-IGNoZWNrbGlzdD88YnI-ZmE6ZmEtY29kZTxicj4gZmE6ZmEtY29nc306Ojpib3hcbiAgRSAtLT4gS3tNYWNoaW5lPGJyPiBhY3Rpb25hYmxlPGJyPiBjaGVja2xpc3Q_PGJyPmZhOmZhLWNvZGU8YnI-IGZhOmZhLWNvZ3N9Ojo6Ym94XG4gIEkgLS0-IEt7TWFjaGluZTxicj4gYWN0aW9uYWJsZTxicj4gY2hlY2tsaXN0Pzxicj5mYTpmYS1jb2RlPGJyPiBmYTpmYS1jb2dzfTo6OmJveFxuICBLIC0tPiB8RW50aXR5IE1hcHBpbmcgdG8gT250b2xvZ3l8IExbb250b2xvZ3k8YnI-IHRhZ2dlZCA8YnI-cmVxdWlyZW1lbnRzXTo6OmJveFxuXG4gIEsgLS0-IHxFbnRpdHkgRGF0YSBUeXBpbmd8IE1bRGF0YTxicj4gdHlwZWQ8YnI-IHJlcXVpcmVtZW50c106Ojpib3hcbiAgTSAtLT4gfERlZmluaXRpb24gb2YgdmFsdWUgc2V0c3wgTltPbnRvbG9neTxicj4gY29uc3RyYWluZWQ8YnI-IGZhOmZhLWxpbmsgcmVxdWlyZW1lbnRzXTo6OmJveFxuICBLIC0tPiB8Rm9ybWFsaXphdGlvbnwgSltNYWNoaW5lIFJlYWRhYmxlIE1ldGFkYXRhIFByb2ZpbGUgPGJyPmZhOmZhLWNvZGU8YnI-IGZhOmZhLWNvZ3M8YnI-ZmE6ZmEtY29kZV06Ojpib3hcbiAgSiAtLT4gfEltcGxlbWVudGF0aW9ufCBPW1VzZXIgRnJpZW5kbHkgTWV0YWRhdGEgQ29sbGVjdGlvbiA8YnI-IGUuZy4gRm9ybSwgVGFidWxhciB0ZW1wbGF0ZSA8YnI-IGZhOmZhLWZpbGUtZXhjZWwtbyBmYTpmYS1maWxlLWV4Y2VsLW8gZmE6ZmEtZmlsZS1leGNlbC1vPGJyPiBmYTpmYS1ncm91cCBmYTpmYS1ncm91cCBmYTpmYS1ncm91cF06Ojpib3hcbiAgXG4gIGxpbmtTdHlsZSAwLDEsMiwzLDQsNSw2LDcsOCw5LDEwLDExLDEyLDEzLDE0LDE1IHN0cm9rZTojMmE5ZmM5LHN0cm9rZS13aWR0aDoxcHgsY29sb3I6IzJhOWZjOSxmb250LWZhbWlseTphdmVuaXI7XG4gIGNsYXNzRGVmIGJveCBmb250LWZhbWlseTphdmVuaXIsZm9udC1zaXplOjE0cHgsZmlsbDojMmE5ZmM5LHN0cm9rZTojMjIyLGNvbG9yOiNmZmYsc3Ryb2tlLXdpZHRoOjFweFxuXG5cblxuICAiLCJtZXJtYWlkIjp7InRoZW1lIjoibmV1dHJhbCJ9LCJ1cGRhdGVFZGl0b3IiOmZhbHNlfQ)
<!--
[](https://mermaid-js.github.io/mermaid-live-editor/#/edit/eyJjb2RlIjoiZ3JhcGggVERcbiAgQVtEZWZpbmluZyBhIE1ldGFkYXRhIFJlcXVpcmVtZW50IFByb2ZpbGU6IFRyYW5zY3JpcHRvbWljcyBEYXRhXSAtLT4gWlxuICBaKGZhOmZhLXBpZS1jaGFydCBSZXF1aXJlbWVudCAgQW5hbHlzaXMpIC0tPiBXW2ZhOmZhLWZpbGUtdGV4dCBmYTpmYS1iYXJzIExpc3Qgb2YgIFJlcXVpcmVtZW50czogTWluaW1hbCB2cyBSZWNvbW1lbmRlZF1cblxuICBXIC0tPiB8U3VydmV5IFN0YXRlIG9mIHRoZSBBcnR8IEN7ZmE6ZmEtYmlub2N1bGFycyA8YnI-IElzIHRoZXJlPGJyPiBwcmlvciB3b3JrP31cblxuXG4gIEMgLS0-IHxOb3wgRVtmYTpmYS1tYWdpYyBDcmVhdGUgQ2hlY2tsaXN0IDxicj5mYTpmYS1jaGVjay1zcXVhcmU8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLXNxdWFyZS1vPGJyPmZhOmZhLWNoZWNrLXNxdWFyZV1cbiAgQyAtLT4gfFllc3wgRFtNaW5pbXVtIEluZm9ybWF0aW9uIENoZWNrbGlzdF1cbiAgRCAtLT4gR3tFdmFsdWF0aW9uOjxicj4gaXMgaXQgZW5vdWdoP31cbiAgRyAtLT4gfFllc3wgSFtmYTpmYS1yZWN5Y2xlIFJldXNlIENoZWNrbGlzdCA8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLWNoZWNrLXNxdWFyZTxicj5mYTpmYS1zcXVhcmUtbzxicj5mYTpmYS1jaGVjay1zcXVhcmVdXG4gIEcgLS0-IHxOb3wgSVtmYTpmYS1jb2RlLWZvcmsgRXh0ZW5kIENoZWNrbGlzdCA8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlPGJyPmZhOmZhLWNoZWNrLXNxdWFyZTxicj5mYTpmYS1zcXVhcmUtbzxicj5mYTpmYS1jaGVjay1zcXVhcmU8YnI-ZmE6ZmEtY2hlY2stc3F1YXJlXVxuICBIIC0tPiBLe01hY2hpbmU8YnI-IGFjdGlvbmFibGU8YnI-IGNoZWNrbGlzdD88YnI-ZmE6ZmEtY29kZTxicj4gZmE6ZmEtY29nc31cbiAgRSAtLT4gS3tNYWNoaW5lPGJyPiBhY3Rpb25hYmxlPGJyPiBjaGVja2xpc3Q_PGJyPmZhOmZhLWNvZGU8YnI-IGZhOmZhLWNvZ3N9XG4gIEkgLS0-IEt7TWFjaGluZTxicj4gYWN0aW9uYWJsZTxicj4gY2hlY2tsaXN0Pzxicj5mYTpmYS1jb2RlPGJyPiBmYTpmYS1jb2dzfVxuICBLIC0tPiB8RW50aXR5IE1hcHBpbmcgdG8gT250b2xvZ3l8IExbb250b2xvZ3k8YnI-IHRhZ2dlZCA8YnI-cmVxdWlyZW1lbnRzXVxuXG4gIEsgLS0-IHxFbnRpdHkgRGF0YSBUeXBpbmd8IE1bRGF0YTxicj4gdHlwZWQ8YnI-IHJlcXVpcmVtZW50c11cbiAgTSAtLT4gfERlZmluaXRpb24gb2YgdmFsdWUgc2V0c3wgTltPbnRvbG9neTxicj4gY29uc3RyYWluZWQ8YnI-IGZhOmZhLWxpbmsgcmVxdWlyZW1lbnRzXVxuICBLIC0tPiB8Rm9ybWFsaXphdGlvbnwgSltNYWNoaW5lIFJlYWRhYmxlIE1ldGFkYXRhIFByb2ZpbGUgPGJyPmZhOmZhLWNvZGU8YnI-IGZhOmZhLWNvZ3M8YnI-ZmE6ZmEtY29kZV1cbiAgSiAtLT4gfEltcGxlbWVudGF0aW9ufCBPW1VzZXIgRnJpZW5kbHkgTWV0YWRhdGEgQ29sbGVjdGlvbiA8YnI-IGUuZy4gRm9ybSwgVGFidWxhciB0ZW1wbGF0ZSA8YnI-IGZhOmZhLWZpbGUtZXhjZWwtbyBmYTpmYS1maWxlLWV4Y2VsLW8gZmE6ZmEtZmlsZS1leGNlbC1vPGJyPiBmYTpmYS1ncm91cCBmYTpmYS1ncm91cCBmYTpmYS1ncm91cF1cblxuXG5cblxuICAiLCJtZXJtYWlkIjp7InRoZW1lIjoibmV1dHJhbCJ9LCJ1cGRhdGVFZGl0b3IiOmZhbHNlfQ)
-->
<!--__Copied from [slides](https://docs.google.com/presentation/d/1zdGdkCGzL2XFUNf4bUMeOXIEX6I8iBlL4-b5a5oV0F4/edit#slide=id.g752ddb65ea_0_3) and [squad2 issue #75](https://github.com/FAIRplus/FAIRPlus_squad2/issues/75)__-->
___
## Capability & Maturity Table
| Capability | Initial Maturity Level | Final Maturity Level |
| :------------- | :------------- | :------------- |
| Interoperability | minimal | repeatable |
___
## Transcriptomics Data model
Large sections of any transcriptomics data model are not unique to transcriptomics experiments, but are common to a wide range of experiments in the biomedical domain. This includes general project- and sample-level information, which will be briefly covered in this recipe but will also be covered in more depth elsewhere.
Where possible, metadata should be mapped to ontologies to maximise the FAIRness of any dataset. This recipe will suggest possible ontologies for metadata fields where these are available. These lists may however not be exhaustive as new ontologies emerge regularly.
### Existing standards and checklists
A set of well-established standards and minimum metadata checklists exist for various aspects of transcriptomics. They include:
Minimum Information About a Microarray Experiment (MIAME) - MIAME is intended to specify all the information necessary for an unambiguous interpretation of a microarray experiment, and potentially to reproduce it. MIAME defines the content, but not the format for this information. The MIAME standard has been in existence for over 20 years and has been widely adopted across the scientific community. The data models of the major transcriptomics repositories such as ArrayExpress, the Expression Atlas and the Gene Expression Omnibus (GEO) are MIAME-compliant.
Minimum Information about a high-throughput nucleotide SEQuencing Experiment (MINSEQE) - MINSEQE describes the minimum metadata that is needed to enable the unambiguous interpretation and facilitate reproduction of the results of the experiment. By analogy to the MIAME guidelines for microarray experiments, adherence to the MINSEQE guidelines will improve integration of multiple experiments across different modalities, thereby maximising the value of high-throughput research. MINSEQE has been integrated into a number of transcriptomics and sequencing archives.
Functional Annotation of Animal Genomes (FAANG) - FAANG provides a set of orthogonal standards for the capture of well-structured metadata for experiments, samples and analyses in the animal genomics domain. The FAANG standards support the MIAME and MINSEQE guidelines, and aim to convert them to a concrete specification.
Human Cell Atlas Metadata (HCA-Metadata) - the HCA metadata model provides a concrete specification for capturing the metadata for single cell sequencing experiments. It is based on the three standards above, while focusing on the specific requirements of the single cell space.
### Minimum metadata vs desirable metadata
A minimum metadata set constitutes metadata items that always need to be supplied with any experimental data. For validation purposes, these should not be omittable. Desirable or recommended metadata items on the other hand, should be supplied if available, but will not cause a validation failure if absent.
### Common metadata
Common metadata include any information that is not specific to transcriptomics experiments but applies to most experiments in the biomedical domain. They include:
- Project level metadata
- Common sample level metadata such as species, tissue, cell type etc.
#### Suggested metadata fields
The following table contains a non-exhaustive list of suggested minimum metadata fields for biological samples. The collection is based on a range of existing metadata standards, including MIAME, MINSEQE, FAANG and HCA. Fields were included if they occurred in at least two of the standards.
|Metadata field|Required|Comment|
|--------|--------|--------|
|unique ID|required||
|sample type|required|ontology field - eg OBI or EFO|
|species|required|ontolofy field - NCBITaxonomy|
|tissue/organism part|required|ontology field - eg Uberon|
|disease|required|ontology field - eg MONDO or DO|
|sex|required|ontology field - eg PATO|
|development stage|required|ontology field - eg Uberon or Hsadpdv; species dependent|
|collection date|required||
|external accessions|recommended|eg Biosamples, Biostudies|
|strain|recommended|ontology field - eg NCBITaxonomy|
|ancestry/ethnicity|recommended|ontology field - eg HANCESTRO|
|age |recommended||
|age unit|recommended|ontology field - eg UO|
|BMI|recommended||
|treatment category|recommended|ontology field - eg OBI, NCIt or OGMS|
|cell type|recommended|ontology field - eg CL|
|growth conditions|recommended||
|strain|recommended|ontology field - species dependent|
|genetic variation|recommended||
|sample collection technique|recommended|ontology field - eg EFO or OBI|
|phenotype|recommended|ontology field - eg HP or MP; species dependent|
|cell cycle|recommended|ontology field - eg GO|
|cell location|recommended|ontology field - eg GO|
### Assay metadata
Assay-level metadata covers any metadata directly related to the preparation of the biomaterial undergoing the assay and the process of performing the assay. Most, though not all, of this metadata is specific to transcriptomics. Examples include:
- Library information, including what sample the library was originally derived from and whether any replicates are biological or technical
- Process (wet experiment) metadata
- Technology type (e.g. bulk RNA-Seq, scRNA-Seq, CITE-Seq)
- Instrument metadata
- Other assay-specific metadata
- QC information
- Workflow metadata
#### Suggested metadata fields
The following table contains a non-exhaustive list of suggested minimum metadata fields for assays. The collection is based on a range of existing metadata standards, including MIAME, MINSEQE and HCA. This list can and should be further broken down based on specific technologies used, such as microarrays or whole genome sequencing.
|Metadata field|Required|Comment|
|--------|--------|--------|
|unique ID|required||
|experiment type|required|ontology field - eg EFO or OBI|
|extracted nucleic acid/material type|required|ontology field - eg ChEBI or EFO|
|platform|required|ontology field - eg EFO or OBI|
|instrument model|required|ontology field - eg EFO or OBI|
|nucleic acid extraction method|required|ontology field - eg EFO or OBI|
|cDNA library amplication method|required|ontology field - eg EFO or OBI|
|array or sequencing method|required|ontology field - eg EFO or OBI|
|biological or technical replicate|required|boolean or CV|
|end bias|required|standardised field or ontology|
|external accessions|recommended|eg protocols.io, AE|
|assay start time|recommended||
|assay end time|recommended||
|assay duration|recommended||
|array quality|recommended||
|cell quality|recommended||
|chemical compound|recommended|ontology field - eg ChEBI|
|labeling molecule used|recommended|ontology field - eg ChEBI|
|spike-in kit used|recommended||
|cDNA primer|recommended|standardised field or ontology|
|library strandedness|recommended|standardised field or ontology|
|cell quality|recommended|standardised field or ontology|
|cell barcode|recommended||
|UMI barcode|recommended||
### Analysis metadata
Analysis-level metadata includes any metadata related to the files that come out of the experiment, from the sequencing or imaging files generated directly by the machine to files generated during the various stages of processing and analysis, as well as details of any analyses performed. It is very important to always capture versions of software and reference genomes used in order to allow accurate replication of results. Analysis metadata includes
- Type of analysis
- File formats, e.g. BAM, fastq or gene count
- File location e.g. URL
- Summarisation of data, e.g. enrichment analysis
#### Suggested metadata fields
The following table contains a non-exhaustive list of suggested minimum metadata fields for analyses. The collection is based on a range of existing metadata standards, including MIAME, MINSEQE and HCA. This list can and should be further broken down based on the specific analysis type (primary, secondary or tertiary analysis, meta-analysis etc)
|Metadata field|Required|Comment|
|--------|--------|--------|
|analysis type|required|ontology field - eg EFO, OBI or EDAM|
|computational method|required|ontology field - eg EFO or EDAM|
|normalisation strategy|required|ontology field - eg EFO or EDAM|
|file format|required|ontology field - eg EDAM|
|file storage location|required||
|software package|recommended||
|software version|recommended||
|analysis date|recommended||
|read index|recommended||
|read length|recommended||
|assembly type|recommended||
|reference genome version|recommended||
## Ontologies for transcriptomics data
While it is essential that any transcriptomics metadata be annotated with ontology terms wherever possible, there is no absolute set of ontologies that must be used above all others. There is however a consensus set of ontologies and other standardised resources that are commonly used in transcriptomics metadata, including in the main data archives. The most commonly used ontologies and fields they apply to, are listed below. This table represents an absolute minimum of ontology annotations that should be included in a transcriptomics metadata set, for it to be considered as FAIR. Not all fields suggested for ontology annotation in the previous section are repeated here, for this reason.
|Data type|Ontology/Entity sources|Type|Notes|
|--|--|--|--|
|Species |NCBI taxonomy Scientific name + ID|Ontology||
|Tissue |Uberon term and Id|Ontology||
|Cell type| CL term and Id|Ontology||
|Disease|MONDO, DO or MeSH|Ontology|no single solution - options vary depending on resource and individual requirements |
|Phenotype/Trait|HPO (human), MP(other mammals), various others for model organisms (yeast, zebrafish, Xenopus, C. elegans)|Ontology||
|Experiment Type|EFO, OBI|Ontology| e.g. RNASeq, CITESeq etc. - |
|Cell location/cycle| GO|Ontology||
|Developmental stage|HSAPDV/Uberon|Ontology||
|Chemical compound| ChEBI|Ontology||
|Chemical compound| UniChem, SMILE, InChi|Entity||
|Gene/protein|ENSEMBL, NCBI/ENTREZ_GENE, UNIPROT, HGNC ID, INSDC|Entity||
|Metabolites| MetaboLights compound accession, ChEBI|Entity||
|Nucleotide reference sequence|ReqSeq|Entity||
## How to generate a metadata template
Using common transcriptomics metadata standards, in particular the fields listed above as guidance, it is possible to easily define a comprehensive metadata template to capture all the experimental variables to describe any transcriptomics experiment in a FAIR-compliant way. The following steps are intended as a starting point to guide the generation of a metadata template.
### Step 1: Define competency questions
- What are the questions you would like to address with the template?
Without a set of a competency questions, important variables may easily be forgotten. It is equally possible to collect too much metadata, making the resulting metadata model opaque and difficult to navigate. Competency questions serve as a guide to identify the most relevant experimental factors. It is worth having competency questions for several types of data user roles, to reduce blind spots.
### Step 2: Define a Minimal Set Of Metadata (MSOM) according to these questions
- Compile metadata from different sources
- Generate a consolidated view on metadata by merging attributes as far as possible
- Differentiate metadata available for most of the studies from metadata occurring rarely (sparse matrix)
- Identify gaps in the metadata available, for most of the studies comprising data, that is considered important, but has not been captured in the past
- Define a MSOM to be captured in the future from the metadata, that is available for most of the studies and the metadata considered to be important
- Identify available community standards, regarding minimal sets of metadata
- Add metadata attributes from those community standards to the MSOM, if they are not yet included
- Assign cardinality to the MSOM (identify mandatory metadata and how many times the attributes may be reported. Some metadata might not be mandatory, but are still important to capture, if available)
- Identify appropriate ontologies representing your data and establish an application ontology (see recipe 4 of UC3)
- Assign, as far as possible, ontologies to the MSOM and the sparse matrix
### Step 3: Introduce semantics into the template
- Identify most important objects to be represented in the model (e.g. study, sample, treatment, result, etc.)
- Make sure to have an appropriate naming strategy for the objects (e.g. a **NGSstudy** is an **OMICSstudy** is a **Study**; do not call a NGSstudy a Study; make sure the granularity fits your purposes)
- Assign MSOM and sparse matrix attributes to the respective objects
- Identify and introduce relationships among the identified objects (e.g. “an NGSstudy contains samples”, “a result is derived from a sample”)
- Identify dependencies to data not represented as objects at this point in time, but, e.g. as term lists
- Make sure that your model can be expanded subsequently to represent those data as objects, as well
- Integrate the sparse matrix of metadata not contained in the MSOM in the model
### Step 4: Reality check
- Introduce measures allowing identifying errors in reported data according to your model
- Expose your model to actual data delivered by independent colleagues and capture the errors and gaps that occurred
- Identify errors and gaps that are related to the model and not occurring due to errors in the data
- Adjust the model according to these errors and gaps
- Re-iterate the reality check until no more severe errors and gaps are occurring that are relevant for the previously defined competency questions
___
## Authors:
| Name | Affiliation | Orcid | Credit role |
| :------------- | :------------- | :------------- |:------------- |
| Danielle Welter | LCSB, University of Luxembourg| [0000-0003-1058-2668](https://orcid.org/orcid.org/0000-0003-1058-2668) | Writing - Original Draft |
|Karsten Quast|||
| Peter Woollard | GSK | [0000-0002-7654-6902](https://orcid.org/0000-0002-7654-6902) | reviewing |
___
## License:
<a href="https://creativecommons.org/licenses/by/4.0/"><img src="https://mirrors.creativecommons.org/presskit/buttons/80x15/png/by-sa.png" height="20"/></a>