# RNA Synthesis
[BI397 at MBL Lab](https://hackmd.io/@ColbyMBL/index)

> By brewbooks from near Seattle, USA - Grand Prismatic Spring, CC BY-SA 2.0, https://commons.wikimedia.org/w/index.php?curid=51511238
## Design of sgRNAs
1. With your lab partner and instructor, **choose a gene to target** for CRISPR. Ideally, lab groups will chose different genes, with some groups choosing "safe" genes, with a history of successful mosaic gene knockout, and others choosing unproven targets.
2. **Obtain the sequence of your target gene**.
- Open a web browers to https://www.ncbi.nlm.nih.gov/protein/
- Enter the name of your gene and the word "Lepidoptera".
- From the list of responses, choose one that is relatively long, and if possible from a common model organism, such as [*Bombyx mori* (the silkmoth)](https://en.wikipedia.org/wiki/Bombyx_mori), *[Manduca sexta* (the tobacco hornworm moth)](https://en.wikipedia.org/wiki/Manduca_sexta, [*Bicyclus anynana*](https://en.wikipedia.org/wiki/Bicyclus_anynana, or [*Eumeta japonica* (the bagworm moth)](https://www.inaturalist.org/taxa/564044-Eumeta-japonica). Record the GenBank accession number and the species in your notes. Then download the amino acid sequence in FASTA format. This can be done by clicking "Send to:". Choose "File" and "FASTA", and click "Create File". This will download the file to your computer.
- Open the file and look at it. FASTA is a simple format for biological sequence data.
- Save and rename the file with a descriptive names, like `Eumeta japonica EGFR protein.fa`
- Next go to http://www.butterflygenome.org/ and open the BLAST tab.
- Copy and paste the amino acid sequence from your orthologous protein into the search window.
- Check the box "V._cardui_unigenes" at the bottom of the window and click "TBLASTN". The results will list *Vanessa cardui* transcripts with sequence similarity to your search query, ranked by their E-value. This metric is analogous to a p-value in statistics, and lower E-values generally indicate a better sequence match.
- Click on the best match (or matches) to see the alignment. Then click on the link to the right that says "FASTA" to download the *Vanessa cardui* transcript nucleotide sequence (as DNA).
- Open the new file and look at it. Save this file with a descriptive name, like `Vanessa cardui c21619_g1 EGFR.fa`
3. **Select an sgRNA for your gene**
- Open Geneious on your computer. Lab computers should already have Geneious installed. If you're working from a personal laptop, you can download a free 14-day trial from https://www.geneious.com/free-trial/
- If Geneious asks for a license key, go to https://www.geneious.com/academic/ to request one.
- Drag and drop your *V. cardui* transcript sequence into the Geneious window.
- From the "Annotate & Predict" menu, select "Find ORFs" adjust the minimum size and apply only the longest and shortest reasonable ORFs (open reading frames). Save the object.
- From the "Tools" menu, select "Cloning" and "Find CRISPR Sites". The default parameters should be ideal, but feel free to discuss them and try other options. Geneious will annotate the sequence with potential sgRNA target sequences. These are color coded, with green being its strongest recommendations. (Be aware that Geneious will include the PAM sequence in the annotation it highlights.)
- Consider these options, in light of the sgRNA design advice above. Discuss the options with your lab partner and instructor. You can mouse-over the annotations to see details like GC content.
- Choose two sgRNA sequences. In your notes, record the guide sequence (5' to 3'), note the position of the end points in your sequence and the strand containing the sequence.
4. Send your sgRNA target sequences to Dr.A.
- In an email to dave.angelini@colby.edu include the following information:
- your target gene's name,
- the name of the species and GenBank accession you used to find the *V. cardui* transcript,
- the cluster and gene ID number of the *V. cardui* transcript (e.g. c21619_g1)
- the sequences of your chosen sgRNA targets, in order of preference,
- the strand on which these guide sequences are found (forward or reverse),
- and the 5' position of each guide sequence.
- Do not include the PAM in your sequences.
- Also attach the ortholog and *V. cardui* transcript FASTA files to the email.
After this week's lab, Dr.A. will place orders for DNA oligos that you will use in the synthesis of sgRNAs.
## Preparation of sgRNAs
It is possible to make sgRNAs in vitro, using a simple, low cost method. After a guide sequence is designed, it can be flanked by a 5$^\prime$ [T7 RNA polymerase](https://en.wikipedia.org/wiki/T7_RNA_polymerase) promoter sequence (taatacgactcactataggg) and 20-nucleotides of the Cas9 recognition sequence (gttttagagctagaaatagc). This oligo can act as one of two primers in a PCR reaction. The second primer has the complementary sequence to the Cas9 backbone, including the 20-nucleotides in the custom oligo. Therefore the two primers have 20-bp of complementarity and overlap. The two primers are used in a PCR reaction (with no other template). The result is a double-stranded DNA with a 5$^\prime$ T7 promoter sequence and the complete Cas9 sgRNA targeting the selected gene-specific sequence. Finally, this DNA served as a template for in vitro RNA synthesis using T7 RNA polymerase.
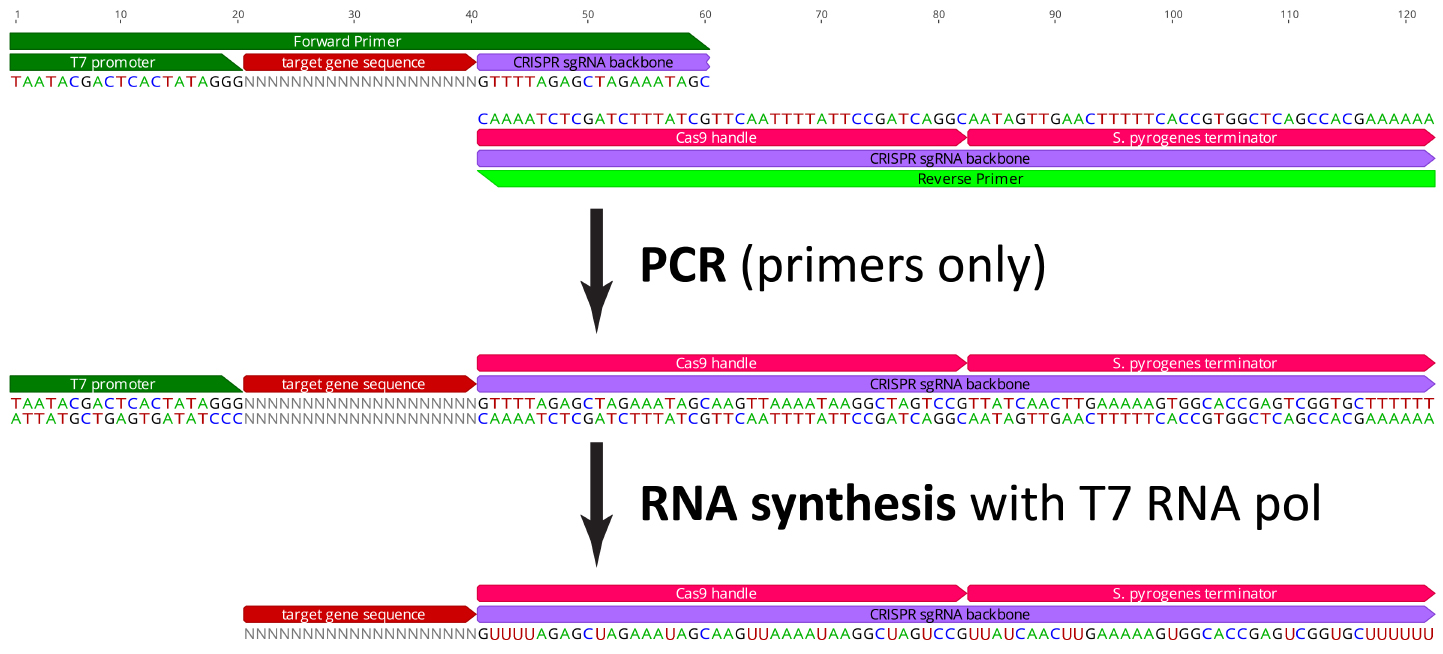
> **Figure 6.** The reagents and steps for synthesis of an sgRNA.
## Working with RNA
Single-strand RNA is prone to degradation by RNase, which can be ubiquitous in the environment. As you prepare RNA, take precautions that will preserve your efforts.
- Keep reagents cold. Let tubes thaw on ice, and keep your reactions on ice until they are ready for incubation at higher temperatures.
- Always use filtered pipette tips to prevent introduction of RNase
- Be sure that water used in your reactions is reagent-grade and nuclease-free.
- Wear clean gloves. Change them if you do something that may expose your hands to RNase, like if you touch a computer, phone or notebook.
- Keep tubes containing your reagents and reactions open as briefly as possible. Don't breathe heavily over them!
## PCR to create a template for an sgRNA
- Combine the following reagents in a 200-μl PCR tube:
| | |
| ------:|:------------------ |
| 6.0 μl | [JumpStart Taq Mix](https://www.sigmaaldrich.com/catalog/product/sigma/d9307) |
| 0.5 μl | gene-specifc primer |
| 0.5 μl | universal-Cas9sg-R primer |
| 5.0 μl | water (nuclease-free) |
| 12.0 μl | total |
- Mix the reaction by pippetting up and down. (Set a pipetter to 3 μl, then gently pipette the solution up and down a few times. Don't intentionally remove any volume.)
- Close the tube and label it.
- Run a thermocycler program with the following conditions. (This should take about 45 minutes.)
| | | |
| ---- |:------ |:------------ |
| 98˚C | 10 s | **\|** |
| 60˚C | 30 s | **\|** 35 cycles |
| 72˚C | 15 s | **\|** |
| 72˚C | 10 min | |
| 12˚C | hold | |
### Quality Control
Confirm the intended PCR product size by running a sample on an agarose gel. There should be a single, strong band.
## In vitro transcription
Next we will use a modification of the MEGAscript T7 Transcription Kit (Life Technologies [item AM1334](https://www.thermofisher.com/order/catalog/product/AM1334)). The nucleotide triphosphates (NTPs) arrive in individual tubes. These should be combined into a new tube with equal volumes (and concentrations) of each.
- Set up the T7 transcription reaction as follows in a 200-μl tube:
| | |
| ----:|:------------------- |
| 7 μl | nuclease-free water |
| 8 μl | NTPs |
| 2 μl | reaction buffer (10X) |
| 1 μl | template PCR product |
| 2 μl | T7 RNA polymerase enzyme mix |
| 20 μl | total |
- Mix the reaction by gently pippetting up and down.
- Incubate the reaction at 37˚C for 1-4 hours, using the incubation mode on the thermocycler. (A range of 1-16 hours seems to work. The optimum appears to be 4 hours.)
- When the incubation is done, without cancelling the run, open the machine and remove the reaction tubes.
- Add 1 μl of [TURBO DNase](https://www.thermofisher.com/order/catalog/product/AM2238), mix well, and return the tubes to incubation at 37˚C for 15 min.
- Cancel the incubation and run the program `Anneal dsRNA`, which denatures the RNA and inactivates the DNase by heating it, then cools slowly to allow complementary RNA to adopt its proper secondary structure.
| | |
| ----:|:------------------- |
| 95˚C | 3 min |
| -0.1˚C/s ramp | takes about 9 min |
| 45˚C | 1 min |
| 12˚C | hold |
- Add 79 μl of nuclease-free water.
- Transfer the solution to a nuclease-free 0.5 ml tube.
## RNA purification
- Add 50 μl of 7.5M ammonium acetate and 300 μl 100% ethanol. Both these reagents should be stored at -20˚C until use.
- Vortex the sample.
- Precipitate the RNA at -20˚C for at least 20 min (or up to 2 days).
- Centrifuge for 20 min at 12,000 rpm at 4˚C. Discard the supernatant.
- Wash the pellet with 0.5 ml cold 70% ethanol.
- Centrifuge for 5 min at 10,000 rpm at 4˚C. Discard the supernatant.
- Dry the pellet in a vacuum centrifuge ("speed-vac") on the medium heat setting. Be sure to leave the caps open to allow evaporation. This should take about 10-15 minutes. Stop before the pellet becomes an opaque white color.
- Resuspend the RNA pellet in 30 μl (suggested) of nuclease-free low Tris-EDTA Buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0)
- Use the NanoDrop spec to measure the concentration and purity of the RNA. Record this information in your notebook. Concentrations should be at least 1 μg/μl, but may vary depending on the length of time allowed for RNA synthesis. Pure RNA should have an A~260~/A~280~ ratio greater than 2.0.
- To confirm the quality of the RNA, run out 1 μl on a 2% agarose gel. It's convenient to run the RNA beside the template PCR product. Double-strand RNA should run very close to DNA of the equivalent sequence and length. However, single-strand RNA will not run to the same distance as double-strand DNA of the same length. Bands much higher than the template DNA represent complex RNA secondary structures that are typically unsuitable for RNAi.
- Based on the measured concentration of RNA, dilute the solution as follows. (Use the old dilution equation, *c~1~v~1~ = c~2~v~2~*. Start with an initial volume, *v~1~*, that estimates the total volume of sgRNA solution available.)
- for sgRNA: 500 ng/μl in nuclease-free low Tris-EDTA buffer
- Aliquot 5 μl into 0.5-ml tubes.
- Store at -80°C.
## Preparation of Cas9/sgRNA solution
### Cas9 prep
#### As described by [Martin et al. 2020](https://jeb.biologists.org/content/223/Suppl_1/jeb208793.long).
- Resuspend 65 μg NLS-Cas9 ([QB3 Berkeley](http://qb3.berkeley.edu/macrolab/cas9-nls-purified-protein/)) in 45 μl RNAse-free water.
- Add 10 μl 0.5% [phenol red](https://en.wikipedia.org/wiki/Phenol_red) solution
- Aliquot 5 μl into 0.5-ml tubes
- Store at -80°C
#### Modified
The NLS-Cas9 arrived in 10 μl aliquots of a solution at 6.4 μg/μl. I diluted this to approximate the concentration specified above using the steps below.
- Add 34.3 μl of nuclease-free water to one Cas9 aliquot.
- Add 9.8 μl of 0.5% phenol red.
- Mix.
- Place 5-μl aliquots in 0.5 ml tubes.
- Store at -80˚C.
#### Combine Cas9 and sgRNA immediately before use
Guide RNAs and Cas9 should be combined just prior to [microinjection](https://hackmd.io/@EcoEvoDevoLab/microinjection).
- Thaw 5 μl of Cas9 stock and 5 μl of sgRNA on ice
- Combine and add 10 μl RNAse-free water
- Mix gently by pipetting
- Incubate at room temperature for about 10 min, then keep on ice until use
Final concentrations should be about 250 ng/μl Cas9, 125 ng/μl sgRNA, and 0.02% phenol red.
---
[BI397 at MBL Lab](https://hackmd.io/@ColbyMBL/index)